Abstract
Significant reprogramming of epigenome is widely described during pathogenesis of breast cancer. Transformation of normal cell to hyperplastic cell and to neoplastic phenotype is associated with aberrant DNA (de)methylation, which, through promoter and enhancer methylation changes, activates oncogenes and silence tumor suppressor genes in variety of tumors including breast. DNA methylation, one of the major epigenetic mechanisms is catalyzed by evolutionarily conserved isoforms namely, DNMT1, DNMT3A and DNMT3B in humans. Over the years, studies have demonstrated intricate and complex regulation of DNMT isoforms at transcriptional, translational and post-translational levels. The recent findings of allosteric regulation of DNMT isoforms and regulation by other interacting chromatin modifying proteins emphasizes functional integrity and their contribution for the development of breast cancer and progression. DNMT isoforms are regulated by several intrinsic and extrinsic parameters. In the present review, we have extensively performed bioinformatics analysis of expression of DNMT isoforms along with their transcriptional and post-transcriptional regulators such as transcription factors, interacting proteins, hormones, cytokines and dietary elements along with their significance during pathogenesis of breast tumors. Our review manuscript provides a comprehensive understanding of key factors regulating DNMT isoforms in breast tumor pathology and documents unsolved issues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
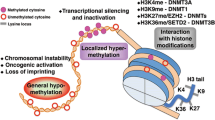
Neoplasia are the uncontrolled growth of cells as a consequence of disrupted gene expression and associated signaling pathways as a consequence of genetic or epigenetic changes (Hanahan and Weinberg 2000). The term ‘epigenetics’ refers to heritable covalent modifications of chromatin components which, by transforming the chromatin organization, affect accessibility of DNA for the regulatory and transcription factors without affecting the basic nucleotide sequence (Egger et al. 2004). The epigenetic machinery regulates gene expression by (a) DNA methylation; (b) post-translational modifications of histones, and (c) non-coding RNAs. Methylation of DNA is a vital process during development, cellular differentiation and tissue homeostasis (Feil and Fraga 2012). DNA methylation is a process, where methyl group is covalently attached to C-5 of the cytosine residue and catalyzed by evolutionarily conserved isoforms of DNA methyl transferases (DNMTs). The mechanism of DNA methylation is widely associated with various physiological processes such as X chromosome inactivation, chromosome stability, genomic imprinting, tissue specific gene expression, repression of transposable elements and aging (Bernstein et al. 2007). Besides various genetic alterations such as mutations, loss of heterozygosity and inducing copy number variations, cancer cells harbor global epigenetic alterations leading to growth and metastasis demonstrating the complex interplay between genetic and epigenetic mechanisms in (dys)regulation of gene expression (Sadikovic et al. 2008). Recent advances in high throughput DNA sequencing and single cell DNA methylation analysis have revealed existence of distinct epigenetic signatures in variety of cancer types and the extent of epigenetic changes is correlated with tumor stage and type (Fernandez et al. 2012). The functions affecting DNMT isoforms including mutations are correlated with the biological characteristics of malignancy and enhance the proliferation, migration, invasion, stemness, epithelial mesenchymal transition and metastasis of tumor cells (Dawson and Kouzarides 2012). The widespread epigenetic defects including DNA methylation in breast tumors instigated us to revisit the regulation of DNMT isoforms in these pathological conditions. Hence, the present review aimed to assimilate the existing knowledge of genetic and epigenetic regulation of DNMT isoforms in breast tumors along with functional consequences.
(Dys)regulation of DNA methylation during tumorigenesis
Based on the structures, DNMT isoforms are classified into DNMT1, DNMT2 and DNMT3 family. DNMT1, DNMT3A and DNMT3B isoforms expressed in human tissues are encoded by distinct genes DNMT1 (DNMT, AIM, MCMT, CXXC9, HSNE1, ADCADN), DNMT3A (DNMT3A2, TBRS, HESJAS), DNMT3B (ICF, ICF1) localized on chromosome 19, 2 and 20 respectively. Three major DNA methyl transferases are involved in initiation and maintenance of DNA methylation patterns in humans: (a) DNMT1 (maintenance methyl transferase), has a strong predilection for hemi-methylated CpG dinucleotides, consequently methylates the newly synthesized DNA strand considering the methylation in the complementary strand as gold standard; (b) DNMT2 is shown to methylate tRNA anticodon loop and DNA methylation activity of DNMT2 is reported to be low or absent; (c) DNMT3 (de novo methyl transferases) isoforms are involved in de novo methylation and non-CpG methylation (Ramsahoye et al. 2000; Jones and Baylin 2002; Laird 2003; Hermann et al. 2003; Goll et al. 2006). DNMT3 consists of three subtypes: DNMT3A, DNMT3B and DNMT3-like protein (DNMT3L). DNMT3A and DNMT3B possess catalytic activities and are regulated by DNMT3L (Okano et al. 1999; Hu et al. 2008).
Alteration of DNA methylation pattern is closely associated with the initiation and progression of tumors. Rauscher et al., 2015 showed that the frequent DNA methylation alteration in promoter regions, introns, far upstream regions, LINE-1 and satellite 2 DNA repeats were associated with the breast cancer development (Rauscher et al. 2015). Increased methylation in promoter CpG islands of specifically tumor suppressor genes including p16INK4A, p15INK4A, p53, p73, TIMP-3, BRCA1, PLCD1, PCDH17, RASSF1A, HIN-1, FOXD3, MLH1, MSH2, ERCC1, RUNX3, GATA-4 and GATA-5 are frequently reported in several cancers such as hematological malignancies and tumors of lung, colon, breast, neurological, liver, nasopharyngeal, ovarian and endometrium (Kang et al. 2001b; Feng et al. 2010; Quintás-Cardama et al. 2012; Xing et al. 2013; Zhu et al. 2015; Cosgrove et al. 2017; Maleva Kostovska et al. 2018; Hentze et al. 2019; Xu et al. 2019). Global hypomethylation of DNA at various genomic locations including CpG-poor promoters, repeat sequences and retrotransposons results in the overexpression of proto-oncogenes and growth factors attributes to hallmarks of cancer. For instance, hypomethylation of uPA resulting in its overexpression is correlated with progression of breast, prostate and brain tumors (Pakneshan et al. 2005; Kandenwein et al. 2011). Several studies have shown that the loss of imprinting of IGF-2 due to hypomethylation leads to uncontrolled proliferation of tumor cells (Leick et al. 2011). Hypomethylation of Alu repeats in the intronic region of TGFB2 and region overlapping the CpG island of the PRDM16 exon has been observed in tumor cell lines (Irizarry et al. 2009). Joo et al. (2018) showed that heritable DNA methylation pattern is a major contributor for the breast cancer risk in multiple case breast cancer families with no known genetic mutation (Joo et al. 2018).
Overexpression of DNMT1, DNMT3A and DNMT3B at both transcriptional and translational levels which, in turn leads to reduced expression of tumor suppressor genes has been reported in several malignancies including colorectal, lung cancer, glioblastomas, hematological malignancies, prostate and breast (De Marzo et al. 1999; Mizuno et al. 2001; Girault et al. 2003; McCabe et al. 2005; Lin et al. 2007; Lorente et al. 2009; Gravina et al. 2013; Yu et al. 2015; San José-Enériz et al. 2017). Enhanced expression of DNMT1 in tumor tissues is a suggestive for increased aggressiveness of the disease and poor prognosis. Rhee et al. (2002) showed that disruption of either DNMT1 or DNMT3B resulted in partial methylation and simultaneous disruption of DNMT1 and DNMT3B resulted in global hypomethylation and reactivation of tumor suppressor genes in both in vitro and in vivo colorectal cancer models leading to reduced proliferation and tumor growth (Rhee et al. 2002). This indicated that coordinated activity of DNMT1 and DNMT3B might be essential for neoplastic transformation. Furthermore, Xiong et al. (2005) demonstrated that endometrioid cancers frequently showing hypermethylation in the promoters of tumor suppressor genes over expressed DNMT1 and DNMT3B and serous endometrial cancers developed due to P53 mutation, loss of heterozygosity and aneuploidy showed substantial reduction in DNMT1 and DNMT3B levels than controls (Xiong et al. 2005). Furthermore, authors also showed that levels of DNMT1 and DNMT3B were higher in poorly differentiated tumorigenic cell lines such as AN3, KLE, RL-95, HEC1A and HEC1B- compared to differentiated non-tumorigenic Ishikawa cell lines (Xiong et al. 2005). Approximately, 30% of breast cancer patients showed overexpression of DNMT3B and 3–5% showed overexpression of DNMT1 and DNMT3A. Roll et al. (2008) showed that over expression of DNMT3B in breast cancer was strongly correlated with total DNMT1 activity (Roll et al. 2008). Furthermore, studies have shown that global DNA methylation and promoter CpG hypermethylation have been reported to occur simultaneously as independent mechanisms during breast tumorigenesis and at various cancer stages. However, it has been proposed that global DNA hypomethylation may be a process to occur at the later stages since increased degree of global hypomethylation of DNA has been noted with increase in lesion progression. On the other hand, promoter hypermethylation may be an early event during breast tumor development (Tan et al. 2013). Taken together, these data suggested that expression of DNMTs is highly regulated in the tissue and regulators of DNMT expression and activity might play an important role in the dysfunction of DNA methylation machinery.
DNA methylation in breast tumors
Paradoxical DNA methylation changes have been observed in breast cancer: regional hypermethylation of specific genes and global hypomethylation. Regional hypermethylation silences genes involved in cell cycle and growth regulation leading to uncontrolled growth of cells, whereas hypomethylation is a requisite for metastasis (Steeg et al. 2003). The semiquantitative methylation changes through mass spectrometry-based analysis and CpG microarray data have shown that regional hypermethylation signatures in breast cancer have unique combination of CpG islands which is correlated with stage of the disease and is proposed for exploring as diagnostic and prognostic marker(s). Several crucial genes such as p16, BRCA1, MLH1, HMSH2, ESR1, ESR2, RARB, CDH9, PRAC2, TDR10, APC, GSTP1, BIN1, BMP6, CST6, DKK3, RASSF1A, HOXD13, SFN, PITX2, SFRP1, CD3D, CD6, LAX1, UBE2C, TOPBP1 and TIMP3 involved in cell cycle, DNA repair and adhesion were hypermethylated in breast tumors (Radpour et al. 2009; Győrffy et al. 2016). Teschendroff et al. (2016) analyzed 397 breast tumor samples including adjacent normal tissue and demonstrated that differential methylation marks which accounted for 20–30% changes in beta values. Furthermore, the authors showed that these epigenetic signatures are heterogenous and epigenetic changes in adjacent stromal cells were responsible for the aggressiveness of tumor progression (Teschendorff et al. 2016).
TCGA-based bioinformatic analysis revealed that expression levels of DNMT1, 3A and 3B altered in several cancers including breast cancers (Fig. 1). Over the years, various independent studies have demonstrated significant role of DNMT isoforms in breast tumors and hence, promoted us to look at changes in expression levels of DNMT isoforms across different types of breast cancers, at different stages and considering menopause status (Fig. 2a). We mined TCGA database using http://ualcan.path.uab.edu/index.html. The expression levels of DNMT1 and DNMT3A showed highest expression levels in triple negative breast cancer patients, where previous studies have observed hypermethylation of tumor suppressor genes. The DNMT1 and DNMT3A levels were down regulated in the fourth stage and interestingly, correlated with global hypomethylation as a marked signature of metastasis. Expression pattern of DNMT isoform did not vary significantly among pre-, peri- and post-menopause status. However, tumor tissues showed significantly increased levels compared to normal tissues. DNMT3B transcripts were low compared to that of DNMT1 and 3A in different types of breast cancers and stages (Fig. 2a). Furthermore, the survival analysis revealed that increased levels of DNMT3B significantly correlated with the decreased overall survival rate (p < 0.01) in breast cancer patients. However, marginal increased levels of DNMT1 (p > 0.05) and DNMT3A (p > 0.05) did not significantly reflected on survival rate (Fig. 2b). This suggested DNMT isoforms and their target genes might serve as good indicators of prognosis in breast cancers and hence we looked at status of various transcriptional regulators of DNMT isoforms in breast tumors.
Differential expression of DNMT isoforms in various cancers. The gene expression of DNMT1, 3A and 3B based on RNA sequence data (transcript per million) for various cancers- breast cancer (normal, n = 114; tumor, n = 1097), glioblastoma (normal, n = 5; tumor, n = 156), thyroid cancer (normal, n = 59; tumor, n = 505), lung cancer (normal, n = 59; tumor, n = 515), cervical cancer (normal, n = 3; tumor, n = 305), colon cancer (normal, n = 41; tumor, n = 286), endometrial cancer (normal, n = 35; tumor, n = 546), head and neck (normal, n = 44; tumor, n = 520), liver cancer (normal, n = 50; tumor, n = 371), melanoma (normal, n = 1; tumor, n = 104), prostate (normal, n = 592; tumor, n = 497), pancreatic (normal, n = 4; tumor, n = 178), renal cancer (normal, n = 72; tumor, n = 533), stomach cancer (normal, n = 34; tumor, n = 415), testis cancer (normal, n = 59; tumor, n = 505), were downloaded from TCGA database and heatmap is plotted
DNMTs levels are altered in breast cancer: Gene expression data from the TCGA was extracted. a The levels of DNMT1, 3A and 3B in major subclasses of breast cancer- normal (n = 114), luminal (n = 566), Her2 positive (n = 37), triple negative (n = 116); different stages of cancer- normal (n = 114), stage 1 (n = 183), stage 2 (n = 615), stage 3 (n = 247), stage 4 (n = 20) and levels in pre- (n = 230), peri- (n = 37) and post- (n = 700) menopausal women are plotted. b Survival analysis for DNMT1, DNMT3A and DNMT3B are carried out and data is shown. Red-High expression level (n = 810), Blue-Low expression level (n = 271)
Status of regulatory proteins influencing expression of DNMT isoforms in breast tumor tissues
FANTOM5 consortium includes single molecule CAGE profiles across 573 human samples covering major mammalian cell steady states. The data contains complete profiles of 250 different cancer cell lines and 152 human post-mortem tissue samples. Zenbu genome browser is a web-based interactive dynamic CAGE and TSS (transcription start site) exploration platform which enables to survey TSS activity within defined genomic region with user selectable alignment (Severin et al. 2014).
Our Zenbu analysis of DNMT isoforms revealed different TSS for single isoforms across the genome with varied activities. The highest and lowest active TSS sites are shown in Fig. 3. The TSS activity of DNMT1 (Fig. 3a) and DNMT3A (Fig. 3b) were highest in triple negative MDA-MB-453 among breast tumor cell lines. Activity of DNMT 3B (Fig. 3c) TSS activity was highest in MCF-7 cell lines compared to MDA-MB-453 cell line. However, the TSS activity of DNMT3L was found nil (0.0) in both breast cancer cell lines (Fig. 3d).
DNMT isoforms locus, TSS and TSS activity in breast cancer. Images are the screen shot of ZENBU browser showing the locus of DNMT1(a), DNMT3A (b), DNMT3B (c) and DNMT3L (d). The track UCSC CpG islands shows number and locus of CpG islands and FANTOM 5 CAGE Phase 1 and 2 track represents histogram of CAGE tag counts across the entire dataset. The last track shows the TSS activity in breast carcinoma cell line MCF 7 and breast carcinoma cell line MDA-MB-453. Purple, antisense strand; Green, sense strand
Transcriptional regulation
Our bioinformatic analysis indicated that DNMT isoforms interact with several transcription factors including p53, SP1, SP3, E2, p300, E47. Transcription factors such as PBX1 and PAX6 were found to interact with only DNMT1, NRSF1, STAT1 with DNMT3A, XBP1 and HFH1 with DNMT3B and SOX5, GFI1, MAX, PPARA with only DNMT3L. However, certain other transcription factors such as HASF2, ATF, MYB, NFKAPPAB, AP4, OLF1, GC, RFX1, IK2, STAF, CREB, E47, EGR1, GATA3, ZID, SREBP1, E2, EGR2, HNF4, HEN1, ELK1, CAP, PAX5, NRF2, AP2, SP1, ARP1, GATA2, E2F, MYOD, AML1, RREB1, P300, GATA1, ARNT, NFE2, NFKB, EGR3, AP1, AHR, LYF1, P53, NGFIC, NMYC, NF1 and MZF1 shown to be bound to all the four DNMT isoforms. The redundancy in binding of these transcription factors is represented in Fig. 4.
Redundancy in transcription factors (TFs) interaction with DNMT isoforms. In vitro experiments such as transcriptional factor arrays, CHIP assays, recombinant DNMT1, 3A, 3B and 3L constructions have revealed that certain TFs can potentially interact with all the isoforms or few isoforms or only with one specific isoform. The TFs binding to − 500 to + 200 region of DNMT isoforms were retrieved from TF-binding input tool and validated using ContraV3 tool and UCSC genome browser. Fluorescent light green, TFs binding only to DNMT1; Purple, TFs binding only with DNMT3B; Pink, TFs interact with DNMT3B; Olive green, TFs bind to DNMT3L only; Overlaps regions shows those TFs which bind to more than one isoform
Transcriptional activation
SP1- and SP3-mediated transcriptional activation of DNMT isoforms
The SP family transcription factors belongs to conserved zinc finger DNA-binding domain proteins that recognize the GC- rich box (GGGCGGG) and GT rich box (GGTGTGGGG). These factors are important for the expression of different housekeeping genes and genes which are deficient of TATA- or CAAT-boxes in their proximal promoters (Hagen et al. 1992). Several SP proteins have been identified (SP1–SP8) and among these SP1 and SP3 are ubiquitously expressed. SP1 is a transcription activator and SP3 acts as either activator or repressor depending upon the context of either promoter region and cell type (Bouwman and Philipsen 2002). Earlier studies have shown that stoichiometric ratio of SP1 and p53 is required for physical interaction with regulatory element for DNMT1 transcription (Lin et al. 2010b). Total of three putative SP1-binding sites identified on DNMT1 promoter region and one among these binding sites (+ 7 to + 20) being proximal to binding site of p53 (+ 30 to + 56). At low levels, SP1 interacts with p53 and represses DNMT1 expression and at higher levels, SP1 targets p53 to proteasomal degradation via MDM2-mediated ubiquitination and directly binds to DNMT1 promoter to initiate transcription (Lin et al. 2010b). The cis-element in DNMT1 promoter located between -147 to -161 was shown activated by SP1 and SP3 independently of each other and p300 was co-activator for SP3-mediated activation (Kishikawa et al. 2002). Studies have also shown that SP1 and SP3 also acts as the transcriptional activators of DNMT3A and DNMT3B. Minimal promoter regions of both DNMT3A and DNMT3B contain SP1-binding site at − 99 to − 87 and − 100 to − 92 respectively. Overexpression of these SP proteins and site directed mutagenesis in the binding sites indicated that DNMT3A and DNMT3B promoter activities are largely dependent on SP1 and SP3-binding sites (Jinawath et al. 2005). TCGA analysis showed that expression of SP1 transcripts were significantly reduced in breast tumor tissues and SP3 RNA levels were significantly decreased in patients with stage IV disease indicating aberrant expression of SP1 and SP3 might be responsible for DNA methylation changes during breast tumorigenesis (Fig. 5).
Transcriptional regulation of DNMT isoforms. Regulation of transcriptional activation and repression of DNMT1, DNMT3A and DNMT3B by different pathways and alteration of these factors in different stages of breast cancer-normal (n = 114), stage 1 (n = 183), stage 2 (n = 615), stage 3 (n = 247), stage 4 (n = 20), the mechanism of deregulation of these factors are shown. SP1 specificity protein 1, SP3 specificity protein, K-RAS Kristen rat sarcoma viral oncoprotein, AP1 Activator protein 1, STAT3 signal transducer and activator of transcription, p300 E1A-binding protein p300, MDM2 Mouse double minute 2, p53 Tumor protein p53, TET1 ten-eleven translocation 1, TET2 10–11 translocation 2, RB, Retinoblastoma susceptibility protein, E2F PRB-binding protein E2F, FOXO3A Forkhead box O3
Involvement of Ras/AP-1 pathway in regulation of DNMT isoforms
Ras superfamily GTPases are the key regulators of cell proliferation, contraction, intracellular asymmetry, cell shape, apoptosis, single and coordinated cell migration. Elevation of Ras-signal has been shown to play an important role in epigenetic silencing of several genes in human tumors (Patra 2008). DNMT1 promoter harbors three c-Jun dependent enhancer regions downstream to P1 and upstream to P2 and P4 and number of AP-1-binding sites in the promoter region which explains the control of Ras signaling pathway on DNMT1 regulation (MacLeod et al. 1995). Aberrant expression of Ras downstream effectors in breast cancer are documented and has been explored as therapeutic target. Pakneshan et al (2005) showed that downregulation of uPA (urokinase type plasminogen activator) in highly metastatic breast cancer cell line MDA-MB-231 via up regulation of Ras-mediated DNMT1 leading to uncontrolled cell growth (Pakneshan et al. 2005). Furthermore, authors showed that the promoter methylation of uPA was reversed in MDA-MB-231 cell lines upon the treatment of 5′-azacytidine (Pakneshan et al. 2005). Elangovan et al (2013) demonstrated that SLC5A8 a putative tumor suppressor gene is inactivated due to promoter hypermethylation via HRas induced expression of DNMT1 leading to tumorigenesis and lung metastasis in murine mammary tumors (Elangovan et al. 2013). Chang et al (2006) demonstrated that induction of Ha-Ras increases promoter methylation of RECK (Reversion inducing cysteine rich protein with Kazal motifs) which was reversed by the addition of 5′-azacytidine and DNMT3B siRNA indicating Ras induced DNMT3B is primarily responsible for the promoter methylation of RECK gene (Chang et al. 2006). Our TCGA analysis showed that 21 missense mutations in RAS gene and 3 missense and 9 truncated mutations in c-JUN. Interestingly, TCGA analysis showed upregulation of RAS in breast tumor tissues and consistency in stage-wise increase was found (Fig. 5).
STAT3-mediated regulation of DNMT isoforms
Signal transducer and activator of transcription 3 (STAT3) belongs to STAT family of transcription factor which upon phosphorylation by the receptor associated tyrosine kinases form homo or heterodimers and translocate into nucleus, where these transcription factors modulate cell proliferation, apoptosis, cell motility, mammary gland involution and angiogenesis (Bromberg and Darnell 2000; Yu et al. 2009). Activation of STAT3 is regulated by phosphorylation at serine and tyrosine residues and post-translationally by the demethylation at K140 and acetylation at K685 (Kang et al. 2015). Constitutive persistent activation of STAT3 has been implicated in the pathogenesis of whole spectrum of malignancies including that of breast tumors (Burke et al. 2001). STAT3 also been shown to increase methylation of CpG islands in genes including PTPN6, ESR1 and SOCS3 via upregulating DNMT1 expression binding to the promoter region (Zhang et al. 2005; Thomas 2012; Huang et al. 2016). Lee et al. (2012) showed that acetylation of STAT3 is crucial for promoter methylation of tumor suppressor genes and treatment with resveratrol resulted in demethylation in breast tumor and melanoma cell lines (Lee et al. 2012). This indicated, role of resveratrol as epigenetic modifier in breast tumors. Elevated levels of acetylation of STAT3 at K685 is known in subjects with melanoma, colon carcinoma and triple negative breast cancer compared to respective normal tissues. STAT3 K685 acetylation led to hypermethylation and silenced several genes including CDKN2, STAT1 and DLEC1 and authors further showed that promoter methylation are not as a consequence of STAT3 phosphorylation at Y705 but due to acetylation (Lee et al. 2012; Thomas 2012). In addition, chromatin immunoprecipitation confirmed the binding of acetylated STAT3 to DNMT1 promoter as a consequence of increased p300 levels and subsequent interaction with DNMT1 in malignant T lymphocytes and breast cancer cell lines (Macaluso et al. 2003; Zhang et al. 2005). TCGA data analysis revealed the significant aberrant expression of both STAT3 and p300 in breast tumor tissues indicating their crucial role in epigenetic changes during tumorigenesis and metastasis (Fig. 5).
MDM2-mediated regulation of DNMT isoforms
MDM2 is a nuclear localized E3 ubiquitin ligase and promotes accelerated cell growth and tumor formation upon inducing proteasomal degradation of tumor suppressor proteins such as TP53 and RB (Michael and Oren 2003; Sdek et al. 2005). Overexpression of MDM2 has been observed in several cancer types including breast tumors. Distinct promoter usage and alternative splicing of MDM2 has been reported in breast cancer cell lines and breast tumor tissues leading to aberrant expression of MDM2 disrupting TP53 pathway in breast tumors (Lukas et al. 2001; Okumura et al. 2002). TCGA analysis also confirmed aberrant expression of MDM2 and p53 in breast tumor tissues indicating their role in breast tumor initiation and progression (Fig. 5).
Transcriptional repression
Role of TP53 in regulating DNMT isoforms
TP53 gene encodes for the tumor suppressor protein containing DNA binding, oligomerization and transcriptional activation domains. In response to cellular stress, TP53 regulates expression of target genes thereby inducing cell cycle arrest, programmed cell death, senescence, DNA repair and metabolic changes (Hager and Gu 2014; Kang et al. 2015; Kruiswijk et al. 2015). Mutations in TP53 gene are associated with variety of malignancies including Li-Fraumeni syndrome (Petitjean et al. 2007), colon cancer (Munro et al. 2005), lung cancer (Peifer et al. 2012), esophageal cancer (Makino et al. 2010), ovarian cancer (Ahmed et al. 2010), breast cancer (Olivier et al. 2006) and are attributed to aggressiveness of the disease (Schmitt et al. 2002). Miller et al. (2005) have shown that TP53 expression signature is consistently associated with patient survival and is a prognostic and predictive indicator in breast cancer (Miller et al. 2005). The loss of TP53 gene is often through large deletions, frame shift mutations, however, many mutations in the tumor cells are found to be single nucleotide missense variants leading to dominant negative phenotype of variable degree. Majority of these mutations are localized to DNA-binding domain resulting in loss of transcriptional function of TP53 (Miller et al. 2005). TP53-binding sites have been identified in the 5′ flanking region and exon-1 (− 19 to + 317) of promoter region of the human DNMT1 gene. Several p53-binding regions were also identified in the 5′ region of the mouse DNMT1 (Peterson et al. 2003; Lin et al. 2010b). In MCF-7 cells overexpression of TP53 showed reduced levels of both SP1 and DNMT1. Coimmunoprecipitation assay showed that TP53 does not bind directly to SP1 and instead promoter activity was reduced with mutant SP1-binding site in luciferase reporter assays indicating that DNMT1 expression is regulated by TP53 via SP1 in breast tumor cells (Zhang et al. 2016). Furthermore, in MDA-MB-468, triple negative basal type breast cancer cell line mutant TP53 was shown to stabilize the DNMT1-MeCP2-HDAC1 complex leading to suppression of ESR1, survivin and cdc25c gene expression via hypermethylation (Estève et al. 2005; Arabsolghar et al. 2013). Under physiological conditions, p53 repressed transcription of both DNMT3A and DNMT3B while inducing TET1 and TET2 which are crucial for the conversion of 5-methyl cytosine to 5-methyl hydroxy cytosine (Laptenko and Prives 2017). In addition, Wang et al. (2005) showed that interaction of DNMT3A with TP53 is crucial for the stability of DNMT3A and transcriptional suppression of TP53-mediated gene expression in MCF-7 cell lines (Wang et al. 2005). TCGA data analysis revealed that deregulation in TP53 expression is significantly correlated with breast cancer stages which may be due to mutations in TP53 gene and/or due to the overexpression of MDM2 which targets TP53 to proteasomal degradation. Mutation analysis for TP53 in TCGA showed 684 missense mutations, 436 truncated mutations, 19 frame shift mutations and 2 fusions have been reported in breast cancer patients (Fig. 5).
RB-mediated regulation of DNMT isoforms
RB gene encodes for the retinoblastoma (RB) protein which negatively regulates cell cycle progression (Weinberg 1995). The protein maintains the overall integrity of the chromatin structure through the interaction with BRG1 SUV39H1 (Shao and Robbins 1995), SWI/SNF (Zhang et al. 2000) and HDAC1 (Luo et al. 1998). The active dephosphorylated form of the protein binds directly to E2F1 promoter region and acts as the transcriptional repression of E2F1 targeted genes and when phosphorylated by CDK3/cyclin-C it promotes G0-G1 transition and progression of the cell cycle (Ren and Rollins 2004). Robertson et al. (2000) demonstrated that DNMT1 forms the complex with RB/E2F/HDAC1 and represses transcription of E2F responsive promoters in both in vitro and in vivo using calf brain (Robertson et al. 2000). DNMT1 promoters in prostate epithelial cell line of both mouse and human harbors functional E2F-binding sites which is crucial for the regulation of RB/E2F (McCabe et al. 2005). Disruption of p16INK4A, maintain RB in its active form, transcription was associated with aberrant CpG DNA methylation in breast cancer cell lines and primary breast tumors (Herman et al. 1995). Macaluso et al. (2003) showed that pRb2/p130-E2F4/5-HDAC1-DNMT1-SUV39H1 multimeric complex suppressed ERα expression by promoter hypermethylation in breast cancer cell lines (Macaluso et al. 2003). In addition, the same laboratory demonstrated that 5′-Aza-2′-deoxycytidine reorganization of pRB2/DNMT1 multimeric complex on ERα gene promoter and modulated its expression in breast cancer cell lines (Macaluso et al. 2007). These data together suggests that RB pathway is crucial for the regulation of DNMT1-mediated gene promoter methylation.
The promoter region of DNMT3A harbors E2F1-binding sites and was transcriptionally repressed by RB/E2F complex formed at these sites. Tang et al. (2012) reported that RB depletion resulted in overexpression of MDM2 leading to transcriptional activation of DNMT3A which subsequently reduced expression of downstream tumor suppressor genes via promoter hypermethylation (Tang et al. 2012). TCGA data analysis revealed that significant alteration in the transcript expression of both RB and E2F can be attributed to mutations in breast tumors- 20 missense mutations, 61 truncated and 2 fusion of RB; 12 missense and 7 truncated mutations in E2F1 (Fig. 5).
FOXO3A-mediated regulation of DNMT isoforms
Forkhead Box O3A (FOXO3A) belongs to forkhead family of transcription factors characterized by the conserved distinct DNA-binding domain ‘forkhead’ (Benayoun et al. 2011). FOXO family proteins have been considered as tumor suppressors due to their inhibitory action on cell proliferation and inducers of apoptosis (Wang et al. 2014). Ectopic overexpression of FOXO3A upregulated Bcl2 interacting mediator of cell death (BIM) resulted in impaired tumor progression in both in vitro and xenograft models of breast tumors (Zou et al. 2008; Smit et al. 2015). In paclitaxel sensitive breast cancer cell lines, paclitaxel reduced tumor cell survival and induced apoptosis by upregulating BIM via FOXO3A (Sunters et al. 2006). Human primary breast tumors negative for phospho-Akt showed FOXO3A in the cytoplasm and high levels of IκB kinase β-modulator of NFκB pro inflammatory pathway. Over expression of FOXO3A reversed the IκB kinase β dependent stimulation of cell cycle progression, proliferation and tumorigenesis in mice (Hu et al. 2004). Yang et al (2014) showed that FOXO3A binds to promoter region (+ 163- + 173) of DNMT3B and negatively regulates promoter activity. FOXO3A nuclear localization reduced the DNMT3B expression by establishing repressed chromatin structure, whereas knockdown of FOXO3A resulted in open chromatin structure and increased DNMT3B mRNA and protein levels (Yang et al. 2014). TCGA analysis showed significantly reduced mRNA expression of FOXO3A in breast tumors which was correlated with stages. Three missense mutations and three fusions are reported in FOXO3A gene according to TCGA-BRCA database (Fig. 5).
Other transcription factors regulating DNMT isoforms
Several other transcription factors have been reported regulating expression of DNMT isoforms in both physiological and pathological conditions. Homeobox B3 induced DNMT3B overexpression resulted in the epigenetic silencing of tumor suppressor gene RASSF1A in MDA-MB-231 cell lines (Palakurthy et al. 2009). The t(8;21) translocation in acute myeloid leukemia induced the formation of RUNX1 (runt-related transcription factor 1)-MTG8 which was shown to interact directly or indirectly with DNMT1 and there by silencing target gene expression (Liu et al. 2005). In breast cancer cells knockdown of RUNX1 resulted in aberrant expression genes related to chromatin organization- NEAT1, MALAT1 and ECM components including fibronectin 1 and fibrillin 2 (Barutcu et al. 2016). In addition, specificity in the expression of transcription factors confined to specific tissue and/or cell type may limit their role in regulating DNA methylation to specific tissues and/or cell types.
The transcription factor screening using TF-binding input showed DNMT isoforms shows redundancy for transcription factors. This indicates the intricate regulation of DNMT isoforms expression (Fig. 4).
Epigenetic regulation of DNMT isoforms
Next, we screened for CpG density on promoters of genes encoding human DNMT isoforms. Our bioinformatic analysis and literature survey indicates that DNMT isoforms contain CpG islands (Fig. 3). However, the complete regulation of DNMT isoforms expression via their CpG sequence methylation is not well understood.
Post-transcriptional regulation of DNMT isoforms
miRNA-mediated regulation of DNMT isoforms
Micro RNAs (miRNAs) induce translational repression, deadenylation or degradation by imperfectly aligning with the 3′ UTR region of target mRNAs (Filipowicz et al. 2008). Dysregulation of miRNA expression have been reported in several tumors including lung, bladder, pancreatic, liver, esophageal, colon, prostate, ovarian and breast (Lu et al. 2005; Melo and Esteller 2011; Ferreira and Esteller 2018). Recent studies have indicated that specific miRNAs regulate DNA methylation machinery and are linked to aberrant methylation pattern in altering cancer epigenome (Fig. 6). The expression of miR-29a and miR-29b were found to target DNMT3A and DNMT3B in in vitro and in vivo models of breast cancer (Sandhu et al. 2012). Knock down and re-expression of miRNAs showed that miR-26b, miR-29a, miR-29b, miR-29c and miR-148b down regulate DNMT3B in breast cancer cells (Sandhu et al. 2012). The miR-143 was shown to directly target DNMT3A mRNA and downregulate protein expression in colorectal cancers (Ng et al. 2009) and breast (Ng et al. 2014). Additionally, miR-194 was demonstrated to regulate DNMT3A expression pattern in drug resistant breast cancer cells and DNMT3A was shown to be the direct target of miR-194 (Le et al. 2010).
Post-transcriptional regulation of human DNA methyl transferases. a miRNAs and HuR protein that target DNMT isoforms are indicated. miRNAs (micro RNAs) bind to either 3′ UTR or coding region destabilizes the mRNA and subsequently protein levels are reduced. HuR (Hu-antigen R) binds to 3′ UTR od DNMT3B and increases its stability. DNMT DNA methyl transferase, miRNA microRNA, mRNA messenger RNA, UTR untranslated region. b The expression levels of these miRNAs in different stages- normal (n = 76), stage 1 (n = 135), stage 2 (n = 427), stage 3 (n = 171), stage 4 (n = 8) of breast cancer are shown
Long noncoding RNA-mediated regulation of DNMT isoforms
Long noncoding RNA (lncRNA) is a pivotal factor in regulating chromatin structure, chromosome looping, nucleosome positioning, DNA methylation and histone modifications (Böhmdorfer and Wierzbicki 2015; Ferreira and Esteller 2018). Numerous studies have demonstrated that lncRNA breast cancer growth (Shen et al. 2015), proliferation, invasion (Shi et al. 2015), apoptosis (Tuo et al. 2015) and chemotherapeutic resistance (Li et al. 2015). Furthermore, Wu et al. (2018) demonstrated that linc00152 promotes tumorigenesis of triple negative breast cancer by targeting DNMT1, DNMT3A and DNMT3B, which resulted in modulation of BRCA1 and PTEN expression both in vitro and in vivo. In addition, authors showed that knockdown of lnc00152 in MDA-MB-231 cells resulted in down regulation of DNMT1, DNMT3A and DNMT3B in association with up regulation of BRCA1 and PTEN leading to decreased proliferation, invasion and enhanced apoptosis of these cells (Wu et al. 2018).
piRNA-mediated regulation
Aberrant expression of piRNAs and piwi family proteins is associated with hall marks of cancer and have shown promise as novel diagnostic and prognostic biomarkers in several malignancies such as lung squamous cell carcinoma, gastric carcinoma, colon adenocarcinoma and breast cancers (Cheng et al. 2011; Mei et al. 2013). Mouse germ cells that were deficient in Piwi subfamily members Mili or Miwi-2 showed defective de novo methylation of transposons (Kuramochi-Miyagawa et al. 2008). Genome wide methylation microarray analysis using HumanMethylation 450 array platform showed MCF cell lines transfected with piRNA mimics showed 117 genes were differentially methylated. Authors validated that mRNA expression of 6 genes -CDK4, FAM150A, KDM3A, LHX5, SYCE1 and VAMP3- were significantly associated with the expression of piRNA (Fu et al. 2014). The direct interaction between piRNA or Piwi family proteins with DNMT3A and DNMT3B have not been explored so far.
HuR (Hu-Antigen R)-mediated post-transcriptional stabilization of DNMT transcripts
HuR protein is a member of embryonic vision family (ELAV) and possess three RNA recognition motifs rich in AU- and U-rich sequences and binds to target mRNAs with higher specificity and affinity. HuR protein is reported to alter the stability of mRNA or translation or both there by regulating target gene expression (Kuwano et al. 2008). HuR has been shown to play major role in cell proliferation, immune response, stress response, senescence and tumorigenesis post-transcriptionally by influencing the stabilization of mRNAs including those of cyclin A, cyclin B1, cyclin D1, c-fos, c-myc, TNF- α, Mcl1, cyclooxygenase-2, β-catenin, p21, p27, p53, VEGF, iNOS, GM-CSF, SIRT1, uPA and uPAR (Levy et al. 1998; Wang et al. 2000b, a; Brennan and Steitz 2001; Ming et al. 2001; Chen et al. 2002; Sengupta et al. 2003; Tran et al. 2003; Lal et al. 2004; Song et al. 2005; Abdelmohsen et al. 2007, 2008). Bioinformatic analysis showed that DNMT3B mRNA 3′ UTR region has a consensus motif HuR and is one of the putative target of HuR. López de Silanes et al. (2009) experimentally showed that HuR bind to DNMT3B mRNA and enhance its stability leading to increased steady state levels of DNMT3B. Furthermore, the authors demonstrated that cisplatin treatment lower DNMT3B levels via inducing the dissociation of DNMT3B mRNA from HuR followed by instability of mRNA in colorectal carcinoma cell lines (López de Silanes et al. 2009). Recently, CRISPR/Cas9-mediated deletion of RMST (rhabdomyosarcoma 2-associated transcript) in MCF cell lines showed that RMST promotes HuR binding to DNMT3B 3′ UTR region increasing stability of DNMT3B and its upregulation (Peng et al. 2020).
Post-translational regulation of DNMT isoforms
Post-translational modifications (PTMs) including phosphorylation, acetylation, SUMOylation, glycosylation, ubiquitination, nitrosylation, sulfation, butyrylation, propionylation, ADP-ribosylation, methylation and citrullination of proteins play significant role in regulating gene expression, protein activity and function (Reinders and Sickmann 2007). Biochemical and molecular biology studies have confirmed that stability, catalytic properties and functions of DNMTs are also regulated by phosphorylation, acetylation, methylation, SUMOylation and ubiquitination (Fig. 7).
Post-translational regulation of human DNA methyl transferases. Summary of covalent post-translational modifications of DNMT1, DNMT3A and DNMT3B are shown along with the position, amino acid modified and biological significance. These modifications include phosphorylation (P), SUMOylation (S), methylation (M), acetylation (A) and ubiquitination (Ub). The proteins mediate these modifications, if any, are indicated. DNMT DNA methyltransferase, SUMO small ubiquitin like modifier, Set7/9 SET domain containing protein 7/9; Set8 SET domain containing protein 8, LSD1 Lysine specific demethylase 1, Tip60 Tat interacting protein 60 kDa; HDAC histone deacetylase, SIRT1 Sirtuin 1, HAUSP herpes virus associated ubiquitin specific protease
Protein phosphorylation
Following the initial identification of insect DNMT1 phosphorylation at S515 (Glickman et al. 1997), enumerable phosphorylated serine and threonine residues are identified in purified DNMT1 from human cells by targeted high-throughput proteomic approaches. More than sixty phosphorylation sites have been mapped on human and mouse DNMT1 protein (https://www.phosphosite.org) albeit only few of them have been functionally characterized. The phosphorylated S515 located within the amino terminal of replication foci targeting domain is require to preserve the interaction between DNMT1 N-terminal and catalytic domains that is crucial for the enzyme activity (Goyal et al. 2007). Casein kinase 1 delta/epsilon reduces DNA-binding affinity of DNMT1 by phosphorylating S146 in the N-terminal regulatory domain (Sugiyama et al. 2010). In mouse and human glial cells, Akt and PKC phosphorylate DNMT1 at S127/S143 and S143 respectively and there by control the interaction of DNMT1 with PCNA and UHRF1 (Hervouet et al. 2010). Phosphorylation of S143 residue by Akt1 stabilizes the DNMT1 protein in a cell cycle dependent manner (Estève et al. 2011). Previous studies have shown that Akt inactivates GSK3β (Ser/Thr kinase) resulting in recruitment of E3-ubiquitin ligase βTrCp followed by degradation of downstream target proteins (Sharma et al. 2002; Taketo 2004). Lin et al (2010a, b) demonstrated that GSK3β phosphorylates DNMT1 at S410 and S414 and induce DNMT1 binding to βTrCp leading to its proteasomal degradation (Lin et al. 2010a). In addition, PKC α, βI, βII, δ, γ, η, ζ and μ phosphorylates DNMT1 and phosphorylation of DNMT1 in its N-terminal domain by PKCζ reduces its methyltransferase activity in vitro. Furthermore, phosphorylation of DNMT1 by CDK 1, 2 and 5 at S154 is shown to enhance the protein stability and activity (Hervouet et al. 2010; Lavoie et al. 2011; Lavoie and St-Pierre 2011). Phosphorylation of either single or multiple residues of both serine and threonine reduces the activity of DNMT1 which is shown involved in the regulation of global DNA methylation changes and tumorigenesis in HEK-293 and HeLa cell lines (Lavoie et al. 2011). Using cell lines and extensive bioinformatic analysis, Esteve et al. (2016) showed that 14-3-3 is a reader protein of DNMT1 S143 and interact with phosphorylated DNMT1 inducing aberrant DNA methylation and alter gene expression leading to tumor progression and cell invasion in breast cancer (Estève et al. 2016). Phosphorylation of DNMT3 family proteins and their functions are not well understood. However, experimental evidences suggests that Casein kinase 2 phosphorylating DNMT3A in both mice and humans (Deplus et al. 2014; Richter et al. 2019). This phosphorylation guides DNMT3A to specific sequence of the genome and controls subnuclear partitioning (Deplus et al. 2014). Sacco et al (2016) showed that phosphorylation of DNMT3A at S7 site is associated with glucose response and regulates target gene expression in human pancreatic beta and HEK 293 cell lines (Sacco et al. 2016).
SUMOylation
Small ubiquitin like modifier (SUMO) proteins are capable of covalently and reversibly attached to other proteins in cells. SUMOylation has emerged as critical post-translational mechanism regulating protein stability, sub cellular localization, enzyme activity and protein–protein interactions (Verger et al. 2003). DNMT1 possess more than ten putative SUMOylation sites throughout its primary amino acid sequences (Xue et al. 2006). Using in vitro wild type/mutant cell lines and in vivo knockout models, Lee and Muller (2009) demonstrated that SUMOylation of DNMT1 is mediated by SUMO1 and is crucial for methyl transferase activity of DNMT1 (Lee and Muller 2009). Recently, Borgermann et al. (2019) showed that 5-Aza-2′-deoxycytidine treatment targets DNMT1-DNA crosslinks by enhancing SUMOylation of DNMT1 in Human U2OS cell lines. Authors, further showed that inhibition of SUMOylation by the knockdown of SUMO E2 enzyme UBC9 strongly impaired the DNMT1-DNA adducts and DNA replication (Borgermann et al. 2019). This indicated that SUMOylation of DNMT1 plays critical in the resolution of DNMT1-DNA adducts post-replicatively. Although the complete mechanism and enzyme which aid in SUMOylation of DNMT3A and DNMT3B are unidentified. However, preliminary data suggest that SUMOylation of DNMT3A transforms its ability to interact with HDAC (Kang et al. 2001a; Ling et al. 2004). Furthermore, Seo et al. (2019) demonstrated that mutations at R882 residues in acute myeloid leukemia patients resulted in enhanced SUMOylation by SUMO1 protein leading to weak complex formation between DNMT3A and HDAC. This weaker association of DNMT3A-HDAC complex induces acetylation of H3K27 and overexpression of PD-L1 attributing to escape from immune surveillance and drug resistance (Seo et al. 2019).
Methylation, acetylation and ubiquitination of DNMT isoforms
Lysine methylation is another functionally important reversible post-translational modification of DNMTs. DNMT1 contains over 120 lysine residues and is methylated at multiple sites (Wang et al. 2009). Wang et al. (2009) showed that in DNMT1 K1096 (K1094 in humans) methylation is regulated by Set7/9 and LSD1 (Lysine specific demethylase 1) and affects global DNA methylation in murine embryonic stem cells (Wang et al. 2009). Parallel research by Esteve et al. (2009), showed that in humans Set7 methylate K142 of DNMT1 and knockdown of Set7 led to increased DNMT1 levels (Estève et al. 2009). Studies have shown that methylation at K142 is inhibited by Akt1-mediated phosphorylation of DNMT1 at S143 and the methylation of DNMT1 at K142 is recognized by CRL4 ubiquitin E3 ligase to target DNMT1 for ubiquitin dependent proteasomal degradation (Estève et al. 2011; Leng et al. 2018). Set8 is also found to regulate DNA methylation targeting methylated DNMT1 and methylated UHRF1 to proteasomal degradation which is an opposite action to LSD1-mediated DNMT1 protection (Zhang et al. 2019). Methylation-dependent DNMT1 proteolysis is tightly coordinated with cell cycle regulation in that activity of DNMT1 being highest in S phase. During S phase of the cell cycle, DNMT1 was protected by LSD1 and PHF20L1 via inhibiting the binding of L3MBTL3 to DNMT1. Upon dissociation from PHF20L1 and reduced LSD1, L3MBTL3 is known to bind to methylated DNMT1 leading to proteolysis of DNMT1 in late S and G2 phases (Leng et al. 2018). Furthermore, L3MBTL3- CRL4 complex is also shown to induce proteolysis of methylated E2F1. DNMT1 along with E2F1 forms complex with HDAC1 and RB53 to regulate target gene expression and degradation of both methylated E2F1 and methylated DNMT1 by L3MBTL3- CRL4 complex indicate their highly controlled regulation during cell cycle (Leng et al. 2018; Levy 2019).
G9a (also known as euchromatin histone methyl transferase)-mediated demethylation of DNMT3A at K47 is demonstrated to be recognized by the chromodomain of methyl-H3K9-binding protein MPP8 (M phase phospho protein) forming DNMT3A/H3K9/MPP8 complex which represses de novo methylation. G9a is shown to methylate DNMT1 at K70 but the functional consequences are yet to be determined (Chang et al. 2011).
Agoston et al (2005) demonstrated that deletion of N-terminal 120 amino acids mapped to destruction domain of DNMT1, which is responsible for proper ubiquitination, results in increased protein stability and genomic cytosine hypermethylation in normal human breast epithelial cells (Agoston et al. 2005). Authors further showed that this destruction domain is dysfunctional in MCF-7 breast cancer cell lines compared to normal human breast epithelial cells and is responsible for differential expression of DNMT1 among these cell lines (Agoston et al. 2005). Furthermore, Zhou et al (2008) showed that deletion of 120 amino acids of N-terminal region inhibits LBH589 (clinically relevant HDAC inhibitor)-induced DNMT1 ubiquitination in MDA-MB-231 cells, indicating that impairment in regular ubiquitination leads to genomic hypermethylation in breast cancer cell lines (Zhou et al. 2008).
DNMT isoforms have been shown to destabilize by acetylation. An acetyltransferase Tip60 along with RGS6 and DAMP1 are shown to promote acetylation of DNMT1 at K173, K1113, K1115, K1117 and subsequently leads to ubiquitination by E3 ligase UHRF1 followed by proteasomal degradation during late S phase. Conversely, HAUSP (herpesvirus-associated ubiquitin specific protease) and HDAC1 protected DNMT1 from proteolysis via deubiquitination and deacetylation respectively (Du et al. 2010). In contrast, SIRT1 is shown to physically interact with DNMT1 and deacetylates DNMT1 both in vitro and in vivo. Deacetylation at K1349 and K1415 residues of DNMT1 by SIRT1 has been shown to enhance the methyl transferase activity of enzyme in breast cancer cell lines (Peng et al. 2011). Using the extensive proteomics analysis 12 new acetylated lysine residues have been identified in DNMT1 both in vitro and in vivo and the deacetylation impaired methylase activity and transcription repression (Peng et al. 2011; Kar et al. 2012).
Regulation of enzyme activity of DNMT isoforms
Auto inhibitory mechanism and allosteric regulation
The autoinhibitory mechanism for methylation has been potential target for novel small molecule inhibitors for cancer therapy. Several SAM and DNA competitors including RG108 (Siedlecki et al. 2006; Asgatay et al. 2014), RG119-1 (Rondelet et al. 2017), SGI-1027 (Datta et al. 2009), CM-272 (San José-Enériz et al. 2017), BIX-01924 (Rotili et al. 2014), DC-05 and DC-517 (Chen et al. 2014) have been shown to acts as demethylating agents and antiproliferative in various human malignancies. Recently, Muvarak et al. (2016) showed that Poly (ADP-ribose) polymerase inhibitors enhanced binding of PARP1 and DNMT1 at the DNA damage site inducing cytotoxic effects in the breast cancer xenograft model (Muvarak et al. 2016). Furthermore, using molecular simulation, Xie et al. (2019) demonstrated that SFG (DNMT1 and DNMT3A inhibitor), DG-05 (selective inhibitor of DNMT1) and GSKex1 (selective inhibitor of DNMT3A) inhibitors binds specifically to SAM-binding pocket in particular Val1580/Trp893, Asn1578/Arg891adn Met1169/Val1665 of DNMT1/DNMT3A via van der Waals interaction and inhibit the methylation (Xie et al. 2019). Recently, Krishna et al. (2017) demonstrated that small molecule DNMT1 inhibitors JFD01881, RJC02836, RJC02837 and 5-azacytidine binds to Cys1226 and Glu1266 within SAM-binding pocket and inhibit methylation activity. Authors further showed that these compounds display significant in vitro anti-proliferative activity using MDA-MB-231 breast cancer cell lines (Krishna et al. 2017).
Regulation of DNMTs by interacting factors
Over the years, mounting evidences have reported that the large variety of proteins interact with DNMT isoforms including methyltransferases (both DNA and histone), DNA/chromatin-binding proteins, chromatin modifiers, tumor suppressors, transcriptional activators and cell cycle regulators (Hervouet et al. 2018). These interactions subsequently results in stimulation or inhibition of enzyme activity, increase or decrease the efficiency of the enzyme, guide DNMT isoforms to methylation sites, enable dissociation from target DNA or specific methylation patterns maintenance. The discrepancy between the high processivity of replication (1 nucleotide per ~ 0.035 s) and low methylation turnover rate (70–450 s per methyl group addition) by recombinant DNMT1 in vitro (Jackson and Pombo 1998; Pradhan et al. 1999) suggests that additional mechanisms and proteins are required for interacting machineries to increase the DNMT isoforms activity normal physiological processes (Fig. 8).
Regulation of DNMT isoforms by interacting factors: Regulation of DNMT1 is cell cycle dependent. The recruitment of DNMT1 to replication site is carried out by MeCP2, MBD2/3. The interaction of DNMT1 with HAUSP, HDAC1/2 and PCNA increases the stability and activity of DNMT1. Furthermore, interaction of DNMT1 with AKT1 and CDK/cyclin increases methylation activity. At the G2/M phase acetylation, SUMOylation followed by ubiquitination be various interacting factors including Tip60, UHRF1, RGS6, SIRT1 and UBC9 leads to decrease in DNMT1 stability and proteasomal degradation (a). Similarly, DNMT3A/3B/3L multimeric complex was shown to be recruited by CUL4A/HP1/SUV39H1 complex and the stability was increased by the interaction with MeCP2/MBD2/3 and HDAC. Stability of DNMT3 multimeric complex was shown to be decreased due to the ubiquitination and the complex was later cleared by proteasomal degradation
DNMT1 carries out post-replicative conservation of methylation patterns encompassing complete genome in successive generation by directly interacting with PCNA. The non-obligatory interaction of DNMT1 and PCNA enhances the efficiency of methylation activity by two fold and aids for diverse enzyme kinetics in faithful propagation of methylation information (Iida et al. 2002). DNMT1 has been shown to bind to DNMT3A and DNMT3B suggesting an intricate network between DNMT isoforms for the efficient and non-erroneous methylation of target DNA (Kim et al. 2002). Although DNMT1 is self-capable to recognize and bind hemi methylated CpG sites, interaction with methylated CpG-binding proteins such as MeCP2, UHRF family and MBD2/3 have shown to increase the enzyme efficiency. MeCP2 induces chromatin compaction by binding to DNA and interact with DNMT1 via TRD domain. MeCP2 and MBD2/3 recognizes methylated CpG sites and MBD3 binds to HDAC1 and HDAC2 which ultimately interact with DNMT1 (Tatematsu et al. 2000; Robertson et al. 2000; Kimura and Shiota 2003; Bronner et al. 2007). These interactions suggest the complex mechanism in maintaining hypomethylation and transcriptional repression. This complex also interacts with DMAP1 (DNMT associated protein 1) and transcriptional coregulator DAXX (death domain associated protein) mediating repression which is independent of HDAC (Muromoto et al. 2004). In addition, these interactions enhance the heterochromatin region formation at highly methylated regions. Recent studies have shown that UHRF1 is essential for DNA methylation maintenance and genetic aberration in UHRF1 leads to hypomethylation which was similar to homozygous knock down in embryonic stem cells (Bostick et al. 2007). Throughout the S-phase co-localization and interaction of UHRF1 and DNMT1 leads to preferential binding of these proteins to hemimethylated DNA sequence along with H3K9me3 (Arita et al. 2008). The crystal structure of SET and Ring associated (SRA) domain of UHRF1 in complex with hemimethylated DNA revealed that the 5-methyl cytosine is flipped out of the DNA double helix suggesting DNMT1 preferably not directly bind to hemimethylated DNA rather DNMT1 is recruited by UHRF1 (Avvakumov et al. 2008; Hashimoto et al. 2008; Qian et al. 2008). Studies showed that UHRF2, another protein of UHRF family also interact with DNMT1 and represses epigenetic changes indicating the non-redundant functions of UHRF1 and UHRF2 during the development (Pichler et al. 2011). Nishiyama et al. (2020) demonstrated that replication timing-dependent dual mono ubiquitination of PCNA associated factor 15 (PAF15) via UHRF1 is a prerequisite for chromatin recruitment of DNMT1 and for high fidelity DNA methylation inheritance in mouse embryonic stem cells (Nishiyama et al. 2020). Moreover, UHRF1, UHRF2 and DNMT1 together interact with DNMT3A and DNMT3B exhibiting the complex interplay in establishing methylation patterns and its maintenance (Fatemi et al. 2002; Pichler et al. 2011). In addition, Liu et al (2018) showed that trimethylation of H3K27 by EZH2 leading to the formation of H3K27me3-EZH2-DNMT1 complex formation and hypermethylation of Kibra (wwc1) gene CpG island resulted in epithelial mesenchymal transition of triple breast cancer cell lines (Liu et al. 2018). Besides these, DNMT1 association with transcription factors and regulators such as CFP1 (Butler et al. 2008), SP1 (Estève et al. 2007), SP3 (Estève et al. 2007), STAT3 (Zhang et al. 2005) and NRIP1 (Kiskinis et al. 2007) aid in the regulation of cell signaling.
On contrary to DNMT1, which is mainly recruited in replication foci during S-pahse of cell cycle, DNMT3A and DNMT3B are not concentrated to these foci. During the replication process, DNMT3B interact with human chromosome associated proteins (hCAP) C, E and G and condensing complexes leading to chromosomal condensation indicating DNMT3B dependent methylation is, at least partially independent of DNA replication (Margot et al. 2001; Geiman et al. 2004). DNMT3A/DNMT3B interactions with DNMT3L has been demonstrated in recruiting DNMT3A and DNMT3B on DNA during genomic imprinting. The DNMT3L and DNMT3A forms either dimer or tetramers which results to refolding of DNMT3A leading to increased de novo enzyme activity up to 20-fold (Suetake et al. 2004; Kareta et al. 2006). The recruitment of DNMT3A/DNMT3L complexes was more frequent on Alu sequences imprinted gene promoters and CpG-rich regions (Jia et al. 2007; Glass et al. 2009). Furthermore, interaction of DNMT3 isoforms with LSH (lymphoid specific helicase) increased the processivity of these enzymes and nullification of LSH in embryonic stem cells provoked hypomethylation of repeat elements and decreased expression of specific genes (Myant and Stancheva 2008). DNA de novo methylation is initiated by SUV39H1 and subsequent binding of HP1 leads to recruitment of DNMT3A and/or DNMT3B on the target sequence (Fuks et al. 2003). DNMT3B interaction with SUV39H1 is involved in the pericentric heterochromatin methylation and not in the methylation of centromeric regions. On the other hand interaction of DNMT3B with CENP-C favors methylation of centromeric areas (Gopalakrishnan et al. 2009).
These DNMTs interacting factors were reported to be altered in tumors. In the CUL4A (component of cullin-ring-based E3 ubiquitin protein ligase complex) over expressing tissues such as hepatomas and breast cancer DNMT3B activity was enhanced due to its interaction with CUL4A-NEDD8 resulting in hypermethylation (Shamay et al. 2010). Jin et al. (2010) reported that UHRF1 is overexpressed in BRCA1 hyper methylated breast tumor tissues and overexpression of UHRF1 in breast cancer cell lines led to deacetylation of H3/H4 followed by DNMT1 recruitment on to BRCA1 promoter and hypermethylation (Jin et al. 2010). Dong et al. (2013) reported that elevation in Snail-SUV39H1 complex was in coordination with the elevation in H3K9me3 at the E-cadherin promoter leading to the recruitment of DNMT1 and gene silencing causing enhanced epithelial mesenchymal transition, a function of DNMT1 which is entirely different from the previously known biological function, in basal like breast cancer cell lines (Dong et al. 2013). Furthermore, Duvall-Noelle et al. (2016) showed that LASP-1 (LIM and SH3 protein 1) interaction with UHRF1-DNMT1-Snail1 complex is associated with alteration in epigenetic modifications leading to breast tumor cell migration, local invasion and metastasis (Duvall-Noelle et al. 2016). Recently, Pradhan et al. (2019) demonstrated that the treatment of breast cancer cell lines with the sublethal dosage of hydrogen peroxide induces DNMT1, Snail, Slug and HDAC1 via ERK pathway and induces chromatin remodeling at the E-cadherin promoter. Authors showed that treatment of breast cancer cell lines with 5-aza-deoxycytidine prevented the promoter CpG methylation of E-cadherin and treatment of cells with ERK inhibitor reduced the expression of DNMT1, Slug and snail indicating the synergistic role of histone methylation, deacetylation and methylation-mediated chromatin remodeling during breast tumorigenesis (Pradhan et al. 2019). Table 1 summarizes alterations of interacting factors of the DNMT isoforms in breast tumor tissues as analyzed in TCGA database.
Hormonal regulation of DNMT isoforms
Hormones are essential for the growth, function, maintenance of tissue homeostasis of mammary gland and variation in the levels are known to cause several pathological conditions. The mammary gland undergoes several major changes postnatally in every stages of woman’s life including puberty, pregnancy, lactation and involution. Initially at birth, mammary gland consists of a primary duct and few secondary ducts and resembles that of man. During puberty, dramatic changes occur including significant development of ducts in terms of elongation, branching and accumulation of fat in the adipose tissue of the breast in females. Along with every ovarian cycle, mammary gland undergoes cyclic changes and major differentiation with lobuloalveolar growth occurs from pregnancy to throughout lactation. Post-lactation involution of mammary gland results in regressed ducts and lobuloalveolar structures. In all these stages of mammary gland hormones play major role at the genetic, molecular and epigenetic levels (Rijnkels et al. 2010; Brisken and O’Malley 2010; Macias and Hinck 2012; Holliday et al. 2018). In the gynecological malignancies such as breast carcinoma, ovarian cancers and endometrial adenocarcinoma involvement of hormones and their receptors in the tumor initiation, growth, invasion and metastasis have been reported (Garrett and Quinn 2008; Rice 2010). The role of hormones (if any) in regulating DNMTs in the breast cancer context is discussed below and illustrated in Fig. 9.
Hormonal regulation of human DNA methyl transferases. Expression of DNMT isoformsin mammary gland in response to different hormones is shown. Estrogen (E2) upon binding to estrogen receptor either directly binds to estrogen response elements on DNA or via activating transcription factors such as AP1and STAT3. Growth factors, cytokines and chemokines activates ERK which in turn phosphorylates estrogen receptor independent of estrogen leading to aberrant DNMT isoforms expression. Progesterone binding to its receptor directly binds to DNA or activates ERK and subsequently activates transcription factors leading to decreased expression of DNMT isoforms. DNMT isoforms also showed to possess binding sites to glucocorticoid receptor and expression of mRNA is influenced by glucocorticoids. On the other hand, gonadotropins such as LH and FSH, prolactin, androgens, melatonin and cortisol shown to induce aberrant expression of DNMT1, DNMT3A and DNMT3B when present, however, the pathway through which they act is unknow. Other hormones which significantly influence breast cancer growth, such as βhCG, oxytocin, Serotonin, T3, T4, TSH, PTH, Calcitonin, growth hormone, Catecholamines and insulin have not yet been shown whether or not they affect expression of DNMT isoforms. E2 estrogen, ER estrogen receptor, ERE estrogen response element, RE response element, Src steroid receptor coactivator, PI3K Phosphatidylionosiyol-3-kinase, TFs transcription factors, P4 progesterone, PR progesterone receptor, GC glucocorticoid, GR glucocorticoid receptor, GRE glucocorticoid response element
Estrogen
Epidemiological data suggests that women are at 100 fold higher risk of breast cancer development than men and bilateral oophorectomy before the age 35 years reduces 75% of life time breast cancer incidence (Santen et al. 2007). Enhanced period of exposure to estrogen due to early menarche, late menopause, obesity and high bone density is shown to be associated with the increased risk of breast cancer (Hsieh et al. 1990). Clinical studies have shown that women with high plasma free estradiol levels experience 2.5 fold higher rate of breast cancer over the years than those who have low plasma free estradiol levels (Kaaks et al. 2005; Beattie et al. 2006). Lowering the estrogen by tamoxifen or raloxifene treatment reduced breast cancer incidence by 38% and aromatase inhibitors reduces it by 50–65% in the high risk women (Cuzick et al. 2003). Furthermore, during adjuvant therapy use of aromatase inhibitors or anti-estrogens showed to prevent the development of cancer occurrence in contralateral breast (Fisher et al. 1998; Santen et al. 2010). Effects of estrogen on target cells in the breast are mediated via several mechanisms (Fig. 9). Most widely accepted mechanism utilizes estrogen receptor-mediated transactivation of genes which favor cell proliferation and survival (Liao et al. 2014; Jameera Begam et al. 2017). Another mechanism elucidates genotoxicity of by-products of estrogen metabolism directly damage DNA altering apoptosis, DNA repair and cell cycle regulations resulting in clonal expansion of pre-cancer cells (Lippert et al. 2003). Cheng et al (2008), for the first time showed that exposure to estrogen altered DNA methylation patterns in humans (Cheng et al. 2008) and subsequently Kovalchuk et al. (2007) showed that estrogen induced epigenetic changes occur prior to the tumor initiation in mice models (Kovalchuk et al. 2007). In vitro studies have shown that estradiol treatment increased DNMT1, DNMT3A and DNMT3B levels, activity, binding to the target genes and methylation (Wu et al. 2019). Furthermore, estrogen receptor alpha (ERα) was shown to interact directly with DNMT1 and DNMT3B and recruit them on the target genome to suppress the gene expression (Si et al. 2016). Our bioinformatic analysis revealed that DNMT1, DNMT3A and DNMT3B promoters harbors multiple ERα-binding sites (Fig. 4).
Progesterone
Progesterone acts as pro-proliferative factor for the breast tissues and functions in concert with estrogen and estrogen receptors to induce the expansion of glandular structures during puberty (Brisken and O’Malley 2010). Due to the mode of action and functions, progesterone and progesterone receptors gained constant attention for their emerging role as critical modulators of gynecological cancers including breast cancer (Diep et al. 2015). Progesterone induces proliferation of breast cancer cells by activating Ras/ERK pathway (Migliaccio et al. 1998). The elevated progesterone levels combined with estrogen levels has shown more detrimental effects on breast by guiding cells towards tumorigenesis than either of them alone (Hankinson et al. 2004). Though the progesterone play key role in breast tumor development its effect on DNA methylation in breast is not studied. The ER+/PR+ breast cancer cells have shown differential methylation pattern than ER−/PR− breast cancer cells (Li et al. 2010a; Verde et al. 2018). Furthermore, studies have demonstrated that progesterone receptor regulates methylation and expression of ESR1 (ERα) upon binding to ESR1 promoter sequence (Verde et al. 2018). In addition, progesterone treatment either alone or combined with estrogen showed to downregulate DNMT1, DNMT3A and DNMT3B levels leading to hypomethylation in human endometrial stromal cells (Yamagata et al. 2009). The authors also showed that varied cyclic levels of these hormones during luteal phase and mid secretory phase were associated with differential levels of DNMT isoforms in endometrium (Yamagata et al. 2009).
Gonadotrophins
Gonadotrophin releasing hormone antagonists, that suppress the release of FSH (Follicle Stimulating Hormone) and LH (Luteinizing Hormone), have been shown to be effective in the treatment of breast cancer in fertile women (Robertson and Blamey 2003). Planeix et al (2015) demonstrated that VEGFR2 negative endothelial cells of breast cancer expressed FSHR and these FSHR positive blood vessels extended 2 mm to 5 mm outside the tumor periphery indicating their involvement in vascular remodeling in anti VEGFR2 resistance breast tumors (Planeix et al. 2015). Sanchez et al (2016), showed that the FSHR and LHR are functionally expressed in several breast cancer cell lines and the extent of expression was found to be involved in the modulation of cell migration and invasion via activation of G proteins on the plasma membrane (Sanchez et al. 2016). Furthermore, the inclusion of LH or FSH in the cancer cell growth medium in vitro phosphorylates moesin (actin remodeling protein) and focal adhesion kinase ultimately leading to the formation of molecular bridges between integrins, focal adhesion complexes and actin which enhances cell motility (Sanchez et al. 2016). Uysal et al (2018), showed that administration of FSH and/or LH analogues caused aberrant expression of DNMT1, DNMT3A, DNMT3B and DNMT3L and also affected their subcellular localization in mouse oocytes and embryos (Uysal et al. 2018). LH surge has been demonstrated to hypomethylate the promoter regions of several genes including CYPA11a1 and CYPA19a1 which are involved in the estrogen and progesterone synthesis (Okada et al. 2016) In addition, gonadotropin surge can causes change in methylation pattern indirectly by controlling estrogen and progesterone levels (Okada et al. 2016).
Pregnancy associated hormones
Upregulation of pregnancy associated hormones such as estrogen, progesterone and others are shown to be responsible for transiently increased risk for breast malignancies during pregnancy and post-partum period. Placental production of estrogen, progesterone, placental growth factor, human chorionic gonadotrophin and placental lactogen leading to substantial alteration in the hormonal milieu during pregnancy which influence the proliferation, growth, differentiation and expansion of mammary gland tissues (Ishida et al. 1992; Smith et al. 2001; Cnattingius et al. 2005; Froehlich et al. 2019). However, these hormones act beneficial in certain circumstances, for instance, ER+/PR+ MCF or T47D breast cancer cell lines co-cultured with first trimester placental tissue showed reduction of breast cancer cell numbers and reduced expression of ERα on these cells which is responsible for proliferation (Tartakover-Matalon et al. 2010). Furthermore, expression of ERβ, antagonist for tumor cell proliferation and invasion was found to be two fold higher in parous women than in nulliparous women (Asztalos et al. 2010). An important hormone in maintaining pregnancy, human chorionic gonadotrophin, is found to mammary gland protective and reduces the risk of breast malignancies (Russo and Russo 2011). Placental hCG along with tumor suppressor inhibin downregulates ERα expression by inducing CpG methylation (Russo and Russo 2007). However, the ectopic expression of β-hCG in breast cancer patients has shown to be associated with high grade tumors and poor prognosis (Chang et al. 2005). Ectopically expressed β-hCG exerts anti-apoptotic effect by blocking TGFβ receptors, promotes invasion and migration by down regulating E-cadherin, inducing ERK1/2 and MMP-2 (Wu and Walker 2006; Li et al. 2013b, c). Although the direct link between β-hCG and DNMT isoforms have not been established, the increased β-hCG has shown to be associated with global DNA hypomethylation in the DNA isolated from the serum of pregnant women compared to non-pregnant women (Pauwels et al. 2016).
Glucocorticoids
Glucocorticoids are involved in the development of mammary gland during puberty and pregnancy (Casey and Plaut 2007). The expression of GR is observed in normal breast and all stages of breast cancer tissue with the decline in expression from normal to precancerous lesions and to malignant carcinomas (Teulings and van Gilse 1977; Allegra et al. 1978). Glucocorticoids exerts anti-proliferative and anti-apoptotic activity on breast cancer epithelial cells, at least in part, via modulating transcriptional regulation of genes encoding cell survival pathways such as SGK1 and MKP1/DUSP1 (Mikosz et al. 2001; Wu et al. 2004; Melhem et al. 2009). Furthermore, using triple negative breast cancer mouse xenograft models, Skor et al. (2013) showed that treatment with GR antagonists might be useful in multidrug resistant triple negative GR+ breast cancer cells (Skor et al. 2013). However, recently Obradovik et al. (2019) demonstrated that GR activity is higher in metastatic breast tumor and higher expression of GR induces lung metastasis in mouse xenograft models (Obradović et al. 2019). However, there are no studies related to influence of glucocorticoids on DNMT isoforms. Our bioinformatic analysis showed that DNMT1 promoter harbor putative-binding site for GR (Fig. 4).
Cortisol
The negative impact of increased stress on human health has been well explained and known to increase the possibilities of developing migraines, diabetes, heart attacks, ulcers and malignancies (Cohen et al. 2012). In accordance with this greater than 70% of breast cancer patients showed high levels of serum cortisol levels. Nineteen years follow up survey of 18,932 women conducted by Nilsen et al. (2008) showed women working in highly fast paced jobs with high stress are more prone to develop breast cancer than the women working at slower pace with less stress (Nielsen et al. 2008). The dysregulated cortisol release showed positive correlation with disease progression, increased mortality rate, recurrence and increased fatigue (Sephton et al. 2000; Abercrombie et al. 2004). The role of cortisol in breast epigenetics has not been established. However, Intabli et al (2019) showed that treatment of triple negative breast cancer cell lines MDA-MB-231 and Hs578T with cortisol decreased the expression of DNMT1 leading to hypomethylation of promoter regions of key tumor suppressor genes including DAPK1, AKT1, ABL1, CDKN1A and MGMT (Intabli et al. 2019).
Oxytocin, prolactin, gonadotrophins, androgens, melatonin, serotonin, thyroid and parathyroid hormones, calcitonin, and catecholamines have been shown to participate in etiology of breast tumor etiology and progression. However, their influence on regulation of DNMT isoforms is poorly understood.
Influence of cytokines and growth factors in regulating DNMT isoforms
Inflammation has been attributed as one of the hallmarks of cancers and altered levels of cytokines has been shown to regulate global DNA methylation changes in breast cancer (Fleischer et al. 2014; Fogel et al. 2017). Numerous cytokines have been shown associated with chronic inflammation designated as tumor enabling characteristic drive pathogenic changes in breast tumor microenvironment (Esquivel-Velázquez et al. 2014). However, understanding of the involvement of these cytokines in epigenetic modulation in breast cancers is sparse. As mentioned earlier, activated STAT3 which is a downstream signaling molecule for several cytokines belong to IL-6 cytokine family including IL-6, transcriptionally activates DNMT1 leading to hypermethylation of anti-apoptotic genes. IL-6 via IL-6R/STAT3 pathway regulates DNMT1 expression in tumor cells (Huang et al. 2016). Our recent studies in clinically characterized human breast tumor tissues, we demonstrated IL-6 induced proteasomal degradation of DNMT1 which led to promoter DNA hypomethylation of VEGFR2 promoter and subsequently to disorganized sprout formation in endothelial cells isolated from malignant part of breast tissue (Hegde et al. 2020).
Growth factors such as epidermal growth factor, fibroblast growth factor, vascular endothelial growth factor, insulin like growth factor1 and 2 are known to be proliferative to breast cells and are positively correlated with disease progression, end stage, metastatic spread, poor diagnosis and mortality (Richard et al. 1987; Adams et al. 2000; Dickson et al. 2000; Zhang and Yee 2000). Among these, only IGF 1 has been shown to regulate DNA methylation. IGF 1 binding to IGF 1R leads to downregulation of miR152 which elevates DNMT1 levels and also by activating Akt and subsequent nuclear translocation of GSK3 leading to prevention of proteasomal degradation of DNMT1 in breast tumors. This results in overall changes in the methylation pattern of cells in vivo (Wen et al. 2017). Breast cancer cells which express human epidermal growth receptor 2 (HER 2) which is activated mainly by epidermal growth factor has shown differential methylation patterns than those breast cells which do not express these receptors (Fiegl et al. 2006). However, underlying mechanisms are unknown.
Nutrition and diet influencing expression of DNMT isoforms and significance in breast cancers
Breast cancers are complex multi-genic disorders and gene-nutrient interactions has been shown as major contributor in health management and disease prevention (Freudenheim et al. 1996; Franceschi et al. 1996; Rock and Demark-Wahnefried 2002). Over the years, studies in the context of diet and nutrition have shown that nutrient drive epigenetic changes to alter gene expression, susceptibility to disease including cancer (Anderson et al. 2012; Singh et al. 2014; Andreescu et al. 2018). Many studies indicate that early life nutrition exert imprinting effects on genome which might influence the risk of developing multifactorial chronic diseases in the adulthood (Junien 2006; Dolinoy et al. 2007). Accumulating evidences suggests that dietary intake of nutrition alter expression of genes involved in cell cycle regulation, apoptosis and tumor suppressor genes (Landis‐Piwowar et al. 2008; Li and Tollefsbol 2010).
S-adenosyl methionine (SAM) is a methyl group donor in methylation reactions catalyzed by DNMT isoforms (Feil and Fraga 2012). SAM is synthesized from dietary precursors such as methionine (essential amino acid), folate, choline and betaine. Reduced availability of these dietary nutrients results in reduction of SAM synthesis leading to DNA methylation changes, while increased availability of methyl donors showed enhanced methylation reactions. Further availability of nutrients involved in one carbon metabolism such as folate, cobalamin, riboflavin, pyridoxin and methionine have been demonstrated to alter cancer related DNA methylation (Cheng and Blumenthal 2008; Zeisel 2009; Niculescu and Lupu 2011). Low intakes of cobalamin, riboflavin, niacin, pyridoxine and methionine positively correlated with an increased risk for breast cancer and supplementation of folic acid showed reduced breast cancer risk in premenopausal women (Maruti et al. 2009).
Phytoestrogens, including resveratrol and genistein are known to interact with estrogen receptors and regulate estrogen-responsive genes (Thanos et al. 2006). Genistein is a isoflavone from soybean demonstrated to alter DNA methylation of several genes such as p21, p16INK4A, c-MYC and BMI1 thereby preventing growth of breast cancer cells (Li et al. 2013a). In addition, Xie et al (2014) showed that genistein decreases DNMT1 expression, methyltransferase activity and global DNA methylation in MCF-7 and MDA-MB-231 cell lines (Xie et al. 2014). Our earlier studies in lab showed genistein induced reduction in PEPCK-C expression is via promoter DNA methylation at cytosine + 34, + 45 and + 71 positions in fibroblasts and contrarily, genistein increased expression of PEPCK-C in HepG2 cell lines (Seenappa et al. 2016). We also demonstrated that genistein maintain glucose homeostasis by inducing glycogenolysis in HepG2 cell lines (Seenappa et al. 2016). Bioflavonoids such as catechins of tea and polyphenols of coffee, curcumin (component of turmeric powder), lycopene found in tomatoes, papayas, watermelons and carrots have shown to alter DNA methylation patterns in various normal and cancer cell lines.
Viral infections regulating DNMT isoforms
The involvement of viruses and the viral oncogenes in regulating DNMTs has been described earlier in several tumors including that of breast (Hattori and Ushijima 2016). Epstein Barr virus (EBV) have been shown to be involved in the etiology of various malignancies including head and neck cancers, T-cell lymphoma, Burkitt’s lymphoma, gastric carcinoma and breast cancer (Amarante and Watanabe 2009; Tempera and Lieberman 2014). Tsai et al. (2002) demonstrated that introduction of EBV product LMP1 (latent membrane protein 1) oncoprotein in to MCF-7 breast cancer cell line activated DNMT1, DNMT3A and DNMT3B resulted in the silencing of CDH1 (Tsai et al. 2002). Human immunodeficiency virus type I induces DNMT1 through the response element in the − 1634 to + 71 region leading to the hypermethylation of p16INK4A. Huschtscha et al. (2001) showed that normal human mammary epithelial cells can be immortalized by SV-40 induced transformation (Huschtscha et al. 2001). Furthermore, Hachana et al. (2009) showed that methylation of TIMP3, RASSF1A, SHP1 and BRCA1 were higher in case of patients with SV40 positive than matched normal breast tissues indicating the role of virus in breast cancer progression (Hachana et al. 2009).
DNMT isoforms as therapeutic targets in breast tumors
With the mounting evidence of how DNA methylation orchestrates abnormal gene expression to drive breast tumorigenesis, there is an increasing focus on developing pharmacological interventions for clinical management. Currently two DNMT inhibitors (DNMTi): 5-azacytidine (Vidaza, Celgene) and 5-aza-2′-deoxycytidine or decitabine (Dacogene, Supergen) have been approved by US Food and Drug Administration (FDA) for the treatment of acute myeloid leukemia and high risk myelodysplastic syndrome respectively (Kaminskas et al. 2005a, b). Phase I and II clinical trials investigating the efficacy of demethylating agents in breast cancer yielded promising results (Connolly et al. 2017). Triple negative breast cancers which do not express ER, PR or HER2 receptors are not amenable to conventional therapies. Li et al (2010a, b) demonstrated that treatment of breast cancer cell lines with DNMT inhibitors induced epigenetic reactivation of endogenous estrogen and progesterone receptors (Li et al. 2010b). Furthermore, clinical phase II trials conducted by Connolly et al. (2017) showed that improved efficacy of the treatment when 5-azacytidine was administered along with the hormonal therapy (Connolly et al. 2017). Yu et al (2018) showed that decitabine treatment significantly decreases DNMT protein levels and inhibits tumor growth in triple negative breast cancer xenograft models (Yu et al. 2018). This indicated levels of DNMT isoforms might serve as prognostic marker in triple negative breast cancer patients. The list of DNMT inhibitors along with their outcome in clinical management of breast cancers is given in Table 2.
Conclusion
The regulation of gene expression, activity and recruitment of DNMT isoforms have been tightly regulated by the coordinated functions of transcriptional, post-transcriptional, translational and post-translational events. Several extrinsic and intrinsic factors such as hormones, growth factors, cytokines, vitamins and life style/nutrients have been demonstrated to modulate DNMT isoforms in health and disease. From the previous studies and our bioinformatic analysis confirm that alteration in any of these might lead to tumor initiation, aggressiveness, metastasis and differential response to drugs. Interestingly, alterations in DNMT expression and function have also been attributed in prognosis of breast cancer subjects. The knowledge of the complete regulation of DNMT isoforms is inadequate. Although number of drugs have been explored against DNMT isoforms in several malignancies clinically, failure to reverse methylation changes and/or preventing further changes in gene expression indicates the necessity and importance of understanding the regulation of expressions of these proteins.
References
Abdelmohsen K, Pullmann R, Lal A et al (2007) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 25:543–557. https://doi.org/10.1016/j.molcel.2007.01.011
Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M (2008) Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389:243–255. https://doi.org/10.1515/BC.2008.022
Abercrombie HC, Giese-Davis J, Sephton S et al (2004) Flattened cortisol rhythms in metastatic breast cancer patients. Psychoneuroendocrinology 29:1082–1092. https://doi.org/10.1016/j.psyneuen.2003.11.003
Adams J, Carder PJ, Downey S et al (2000) Vascular endothelial growth factor (VEGF) in breast cancer: comparison of plasma, serum, and tissue VEGF and microvessel density and effects of tamoxifen. Cancer Res 60:2898–2905
Agoston AT, Argani P, Yegnasubramanian S et al (2005) Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem 280:18302–18310. https://doi.org/10.1074/jbc.M501675200
Ahmed AA, Etemadmoghadam D, Temple J et al (2010) Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 221:49–56. https://doi.org/10.1002/path.2696
Allegra JC, Lippman ME, Thompson EB et al (1978) Association between steroid hormone receptors and response rate to cytotoxic chemotherapy in metastatic breast cancer. Cancer Treat Rep 62:1281–1286
Amarante MK, Watanabe MAE (2009) The possible involvement of virus in breast cancer. J Cancer Res Clin Oncol 135:329–337. https://doi.org/10.1007/s00432-008-0511-2
Anderson OS, Sant KE, Dolinoy DC (2012) Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 23:853–859. https://doi.org/10.1016/j.jnutbio.2012.03.003
Andreescu N, Puiu M, Niculescu M (2018) Effects of dietary nutrients on epigenetic changes in cancer. In: Dumitrescu RG, Verma M (eds) Cancer epigenetics for precision medicine: methods and protocols. Springer, New York, pp 121–139
Arabsolghar R, Azimi T, Rasti M (2013) Mutant p53 binds to estrogen receptor negative promoter via DNMT1 and HDAC1 in MDA-MB-468 breast cancer cells. Mol Biol Rep 40:2617–2625. https://doi.org/10.1007/s11033-012-2348-7
Arita K, Ariyoshi M, Tochio H et al (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455:818–821. https://doi.org/10.1038/nature07249
Asgatay S, Champion C, Marloie G et al (2014) Synthesis and evaluation of analogues of N-phthaloyl-l-tryptophan (RG108) as inhibitors of DNA methyltransferase 1. J Med Chem 57:421–434. https://doi.org/10.1021/jm401419p
Asztalos S, Gann PH, Hayes MK et al (2010) Gene expression patterns in the human breast after pregnancy. Cancer Prev Res 3:301–311. https://doi.org/10.1158/1940-6207.CAPR-09-0069
Avvakumov GV, Walker JR, Xue S et al (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455:822–825. https://doi.org/10.1038/nature07273
Barutcu AR, Hong D, Lajoie BR et al (2016) RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta BBA Gene Regul Mech 1859:1389–1397. https://doi.org/10.1016/j.bbagrm.2016.08.003
Beattie MS, Costantino JP, Cummings SR et al (2006) Endogenous sex hormones, breast cancer risk, and tamoxifen response: an ancillary study in the NSABP breast cancer prevention trial (P-1). J Natl Cancer Inst 98:110–115. https://doi.org/10.1093/jnci/djj011
Ben-Baruch N, Denicoff AM, Goldspiel BR et al (1993) Phase II study of fazarabine (NSC 281272) in patients with metastatic colon cancer. Invest New Drugs 11:71–74. https://doi.org/10.1007/BF00873915
Benayoun BA, Caburet S, Veitia RA (2011) Forkhead transcription factors: key players in health and disease. Trends Genet 27:224–232. https://doi.org/10.1016/j.tig.2011.03.003
Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128:669–681. https://doi.org/10.1016/j.cell.2007.01.033
Bimonte S, Cascella M, Barbieri A, et al (2019) Shining a light on the effects of the combination of (–)-epigallocatechin-3-gallate and tapentadol on the growth of human triple-negative breast cancer cells. In Vivo 33:1463–1468. https://doi.org/10.21873/invivo.11625
Böhmdorfer G, Wierzbicki AT (2015) Control of chromatin structure by long noncoding RNA. Trends Cell Biol 25:623–632. https://doi.org/10.1016/j.tcb.2015.07.002
Borgermann N, Ackermann L, Schwertman P et al (2019) SUMOylation promotes protective responses to DNA-protein crosslinks. EMBO J 38:e101496. https://doi.org/10.15252/embj.2019101496
Bostick M, Kim JK, Estève P-O et al (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764. https://doi.org/10.1126/science.1147939
Borges S, Döppler HR, Storz P (2014) A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res Treat 144(1):79–91
Bouwman P, Philipsen S (2002) Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol 195:27–38. https://doi.org/10.1016/S0303-7207(02)00221-6
Braiteh F, Soriano AO, Garcia-Manero G, Hong D, Johnson MM, Silva LDP, Yang H, Alexander S, Wolff J, Kurzrock R (2008) Phase I study of epigenetic modulation with 5-Azacytidine and Valproic Acid in patients with advanced cancers. Clin Cancer Res 14(19):6296–6301
Brennan CM, Steitz JA (2001) HuR and mRNA stability. CMLS Cell Mol Life Sci 58:266–277. https://doi.org/10.1007/PL00000854
Brisken C, O’Malley B (2010) Hormone action in the mammary gland. Cold Spring Harb Perspect Biol 2:a003178. https://doi.org/10.1101/cshperspect.a003178
Bromberg J, Darnell JE (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19:2468–2473. https://doi.org/10.1038/sj.onc.1203476
Bronner C, Achour M, Arima Y et al (2007) The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther 115:419–434. https://doi.org/10.1016/j.pharmthera.2007.06.003
Brueckner B, Rius M, Markelova MR, Fichtner I, Hals P-A, Sandvold ML, Lyko F (2010) Delivery of 5-azacytidine to human cancer cells by elaidic acid esterification increases therapeutic drug efficacy. Mol Cancer Ther 9(5):1256–1264
Burke WM, Jin X, Lin H-J et al (2001) Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene 20:7925–7934. https://doi.org/10.1038/sj.onc.1204990
Butler JS, Lee J-H, Skalnik DG (2008) CFP1 interacts with DNMT1 independently of association with the setd1 histone H3K4 methyltransferase complexes. DNA Cell Biol 27:533–543. https://doi.org/10.1089/dna.2007.0714
Byun H-M, Choi SH, Laird PW, Trinh B, Siddiqui MA, Marquez VE, Yang AS (2008) 2′-Deoxy-N4-[2-(4-nitrophenyl)ethoxycarbonyl]-5-azacytidine: A novel inhibitor of DNA methyltransferase that requires activation by human carboxylesterase 1. Cancer Lett 266(2):238–248
Candelaria M, Gallardo-Rincón D, Arce C, Cetina L, Aguilar-Ponce JL, Arrieta Ó, González-Fierro A, Chávez-Blanco A, de la Cruz-Hernández E, Camargo MF, Trejo-Becerril C, Pérez-Cárdenas E, Pérez-Plasencia C, Taja-Chayeb L, Wegman-Ostrosky T, Revilla-Vazquez A, Dueñas-González A (2007) A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol 18(9):1529–1538
Casey TM, Plaut K (2007) The role of glucocorticoids in secretory activation and milk secretion, a historical perspective. J Mammary Gland Biol Neoplasia 12:293–304. https://doi.org/10.1007/s10911-007-9055-3
Casper ES, Schwartz GK, Kelsen DP (1992) Phase II trial of fazarabine (arabinofuranosyl-5-azacytidine) in patients with advanced pancreatic adenocarcinoma. Invest New Drugs 10:205–209. https://doi.org/10.1007/BF00877247
Chang E, Lee E, Oh SJ et al (2005) The immunoexpressions and prognostic significance of inhibin alpha and beta human chorionic gonadotrophins (hCG) in breast carcinomas. Cancer Res Treat 37:241–246. https://doi.org/10.4143/crt.2005.37.4.241
Chang H-C, Cho C-Y, Hung W-C (2006) Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b–induced promoter methylation. Cancer Res 66:8413–8420. https://doi.org/10.1158/0008-5472.CAN-06-0685
Chang Y, Sun L, Kokura K et al (2011) MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nature Commun 2:1–10. https://doi.org/10.1038/ncomms1549
Chen C-YA, Xu N, Shyu A-B (2002) Highly selective actions of HuR in antagonizing au-rich element-mediated mRNA destabilization. Mol Cell Biol 22:7268–7278. https://doi.org/10.1128/MCB.22.20.7268-7278.2002
Chen S, Wang Y, Zhou W et al (2014) Identifying novel selective non-nucleoside DNA Methyltransferase 1 inhibitors through docking-based virtual screening. J Med Chem 57:9028–9041. https://doi.org/10.1021/jm501134e
Cheng ASL, Culhane AC, Chan MWY et al (2008) Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res 68:1786–1796. https://doi.org/10.1158/0008-5472.CAN-07-5547
Cheng J, Guo J-M, Xiao B-X et al (2011) piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412:1621–1625. https://doi.org/10.1016/j.cca.2011.05.015
Cheng X, Blumenthal RM (2008) Mammalian DNA methyltransferases: a structural perspective. Structure 16:341–350. https://doi.org/10.1016/j.str.2008.01.004
Cnattingius S, Torrång A, Ekbom A et al (2005) Pregnancy aharacteristics and maternal risk of breast cancer. JAMA 294:2474–2480. https://doi.org/10.1001/jama.294.19.2474
Cohen S, Janicki-Deverts D, Doyle WJ et al (2012) Chronic stress, glucocorticoid receptor resistance, inflammation, and disease risk. PNAS 109:5995–5999. https://doi.org/10.1073/pnas.1118355109
Connolly RM, Li H, Jankowitz RC et al (2017) Combination epigenetic therapy in advanced breast cancer with 5-azacitidine and entinostat: a phase II National Cancer Institute/Stand up to cancer study. Clin Cancer Res 23:2691–2701. https://doi.org/10.1158/1078-0432.CCR-16-1729
Cosgrove CM, Cohn DE, Hampel H et al (2017) Epigenetic silencing of MLH1 in endometrial cancers is associated with larger tumor volume, increased rate of lymph node positivity and reduced recurrence-free survival. Gynecol Oncol 146:588–595. https://doi.org/10.1016/j.ygyno.2017.07.003
Cuzick J, Powles T, Veronesi U et al (2003) Overview of the main outcomes in breast-cancer prevention trials. The Lancet 361:296–300. https://doi.org/10.1016/S0140-6736(03)12342-2
Datta J, Ghoshal K, Denny WA et al (2009) A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res 69:4277–4285. https://doi.org/10.1158/0008-5472.CAN-08-3669
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150:12–27. https://doi.org/10.1016/j.cell.2012.06.013
De Marzo AM, Marchi VL, Yang ES et al (1999) Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res 59:3855–3860
Deplus R, Blanchon L, Rajavelu A et al (2014) Regulation of DNA methylation patterns by CK2-mediated phosphorylation of DNMT3a. Cell Rep 8:743–753. https://doi.org/10.1016/j.celrep.2014.06.048
Dickson C, Spencer-Dene B, Dillon C, Fantl V (2000) Tyrosine kinase signalling in breast cancer: fibroblast growth factors and their receptors. Breast Cancer Res 2:191. https://doi.org/10.1186/bcr53
Diep CH, Daniel AR, Mauro LJ et al (2015) Progesterone action in breast, uterine, and ovarian cancers. J Mol Endocrinol 54:R31–R53. https://doi.org/10.1530/JME-14-0252
Dolinoy DC, Weidman JR, Jirtle RL (2007) Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 23:297–307. https://doi.org/10.1016/j.reprotox.2006.08.012
Dong C, Wu Y, Wang Y et al (2013) Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene 32:1351–1362. https://doi.org/10.1038/onc.2012.169
Du Z, Song J, Wang Y et al (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal 3:ra80. https://doi.org/10.1126/scisignal.2001462
Duvall-Noelle N, Karwandyar A, Richmond A, Raman D (2016) LASP-1: a nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene 35:1122–1133. https://doi.org/10.1038/onc.2015.166
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463. https://doi.org/10.1038/nature02625
Elangovan S, Pathania R, Ramachandran S et al (2013) Molecular mechanism of SLC5A8 inactivation in breast cancer. Mol Cell Biol 33:3920–3935. https://doi.org/10.1128/MCB.01702-12
Esquivel-Velázquez M, Ostoa-Saloma P, Palacios-Arreola MI et al (2014) The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res 35:1–16. https://doi.org/10.1089/jir.2014.0026
Estève P-O, Chin HG, Pradhan S (2005) Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. PNAS 102:1000–1005. https://doi.org/10.1073/pnas.0407729102
Estève P-O, Chin HG, Pradhan S (2007) Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J Biol Chem 282:2615–2625. https://doi.org/10.1074/jbc.M606203200
Estève P-O, Chin HG, Benner J et al (2009) Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. PNAS 106:5076–5081. https://doi.org/10.1073/pnas.0810362106
Estève P-O, Chang Y, Samaranayake M et al (2011) A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 18:42–48. https://doi.org/10.1038/nsmb.1939
Estève P-O, Zhang G, Ponnaluri VKC et al (2016) Binding of 14–3-3 reader proteins to phosphorylated DNMT1 facilitates aberrant DNA methylation and gene expression. Nucleic Acids Res 44:1642–1656. https://doi.org/10.1093/nar/gkv1162
Fatemi M, Hermann A, Gowher H, Jeltsch A (2002) Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem 269:4981–4984. https://doi.org/10.1046/j.1432-1033.2002.03198.x
Feil R, Fraga MF (2012) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13:97–109. https://doi.org/10.1038/nrg3142
Feng W, Orlandi R, Zhao N et al (2010) Tumor suppressor genes are frequently methylated in lymph node metastases of breast cancers. BMC Cancer 10:378. https://doi.org/10.1186/1471-2407-10-378
Fernandez AF, Huidobro C, Fraga MF (2012) De novo DNA methyltransferases: oncogenes, tumor suppressors, or both? Trends Genet 28:474–479. https://doi.org/10.1016/j.tig.2012.05.006
Ferreira HJ, Esteller M (2018) Non-coding RNAs, epigenetics, and cancer: tying it all together. Cancer Metastasis Rev 37:55–73. https://doi.org/10.1007/s10555-017-9715-8
Fiegl H, Millinger S, Goebel G et al (2006) Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res 66:29–33. https://doi.org/10.1158/0008-5472.CAN-05-2508
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. https://doi.org/10.1038/nrg2290
Fisher B, Costantino JP, Wickerham DL et al (1998) Tamoxifen for prevention of breast cancer: report of the national surgical adjuvant breast and bowel project p-1 study. J Natl Cancer Inst 90:1371–1388. https://doi.org/10.1093/jnci/90.18.1371
Fleischer T, Frigessi A, Johnson KC et al (2014) Genome-wide DNA methylation profiles in progression to in situand invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol 15:435. https://doi.org/10.1186/s13059-014-0435-x
Fogel O, Richard-Miceli C, Tost J (2017) Chapter six—epigenetic changes in chronic inflammatory diseases. In: Donev R (ed) Advances in protein chemistry and structural biology. Academic Press, New York, pp 139–189
Franceschi S, Favero A, Russo A et al (1996) Intake of macronutrients and risk of breast cancer. The Lancet 347:1351–1356. https://doi.org/10.1016/S0140-6736(96)91008-9
Freudenheim JL, Marshall JR, Vena JE et al (1996) Premenopausal breast cancer risk and intake of vegetables, fruits, and related nutrients. J Natl Cancer Inst 88:340–348. https://doi.org/10.1093/jnci/88.6.340
Froehlich K, Schmidt A, Heger JI et al (2019) Breast cancer, placenta and pregnancy. Eur J Cancer 115:68–78. https://doi.org/10.1016/j.ejca.2019.03.021
Fu A, Jacobs DI, Zhu Y (2014) Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol 11:1301–1312. https://doi.org/10.1080/15476286.2014.996091
Fuks F, Hurd PJ, Deplus R, Kouzarides T (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31:2305–2312. https://doi.org/10.1093/nar/gkg332
Garrett A, Quinn MA (2008) Hormonal therapies and gynaecological cancers. Best Pract Res Clin Obstet Gynaecol 22:407–421. https://doi.org/10.1016/j.bpobgyn.2007.08.003
Geiman TM, Sankpal UT, Robertson AK et al (2004) DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem Biophys Res Commun 318:544–555. https://doi.org/10.1016/j.bbrc.2004.04.058
Girault I, Tozlu S, Lidereau R, Bièche I (2003) Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 9:4415–4422
Glass JL, Fazzari MJ, Ferguson-Smith AC, Greally JM (2009) CG dinucleotide periodicities recognized by the Dnmt3a–Dnmt3L complex are distinctive at retroelements and imprinted domains. Mamm Genome 20:633. https://doi.org/10.1007/s00335-009-9232-3
Glickman JF, Pavlovich JG, Reich NO (1997) Peptide mapping of the murine DNA methyltransferase reveals a major phosphorylation site and the start of translation. J Biol Chem 272:17851–17857. https://doi.org/10.1074/jbc.272.28.17851
Goll MG, Kirpekar F, Maggert KA et al (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398. https://doi.org/10.1126/science.1120976
Gopalakrishnan S, Sullivan BA, Trazzi S et al (2009) DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18:3178–3193. https://doi.org/10.1093/hmg/ddp256
Goyal R, Rathert P, Laser H et al (2007) Phosphorylation of serine-515 activates the mammalian maintenance methyltransferase DNMT1. Epigenetics 2:155–160. https://doi.org/10.4161/epi.2.3.4768
Gravina GL, Ranieri G, Muzi P et al (2013) Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol Rep 29:1189–1195. https://doi.org/10.3892/or.2012.2192
Győrffy B, Bottai G, Fleischer T et al (2016) Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer 138:87–97. https://doi.org/10.1002/ijc.29684
Hachana M, Trimeche M, Ziadi S et al (2009) Evidence for a role of the Simian Virus 40 in human breast carcinomas. Breast Cancer Res Treat 113:43–58. https://doi.org/10.1007/s10549-008-9901-z
Han D, Wu G, Chang C, Zhu F, Xiao Y, Li Q, Zhang T, Zhang L (2015) Disulfiram inhibits TGF-β-induced epithelial-mesenchymal transition and stem-like features in breast cancer via ERK/NF-κB/Snail pathway. Oncotarget 6(38):40907–40919
Hagen G, Müller S, Beato M, Suske G (1992) Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res 20:5519–5525. https://doi.org/10.1093/nar/20.21.5519
Hager KM, Gu W (2014) Understanding the non-canonical pathways involved in p53-mediated tumor suppression. Carcinogenesis 35:740–746. https://doi.org/10.1093/carcin/bgt487
Hanahan D, Weinberg RA (2000) The Hallmarks of cancer. Cell 100:57–70. https://doi.org/10.1016/S0092-8674(00)81683-9
Hankinson SE, Colditz GA, Willett WC (2004) Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res 6:213. https://doi.org/10.1186/bcr921
Hashimoto H, Horton JR, Zhang X et al (2008) The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455:826–829. https://doi.org/10.1038/nature07280
Hattori N, Ushijima T (2016) Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med 8:10. https://doi.org/10.1186/s13073-016-0267-2
Hegde M, Guruprasad KP, Ramachandra L et al (2020) Interleukin-6–mediated epigenetic control of the VEGFR2 gene induces disorganized angiogenesis in human breast tumors. J Biol Chem 295:12086–12098. https://doi.org/10.1074/jbc.RA120.012590
Hentze JL, Høgdall CK, Høgdall EV (2019) Methylation and ovarian cancer: can DNA methylation be of diagnostic use? (Review). Mol Clin Oncol 10:323–330. https://doi.org/10.3892/mco.2019.1800
Herman JG, Merlo A, Mao L et al (1995) Inactivation of the CDKN2/p16/MTS1 gene Is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 55:4525–4530
Hermann A, Schmitt S, Jeltsch A (2003) The human Dnmt2 has residual DNA-(Cytosine-C5) methyltransferase activity. J Biol Chem 278:31717–31721. https://doi.org/10.1074/jbc.M305448200
Hervouet E, Lalier L, Debien E et al (2010) Disruption of DNMT1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS ONE. https://doi.org/10.1371/journal.pone.0011333
Hervouet E, Peixoto P, Delage-Mourroux R et al (2018) Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenet 10:17. https://doi.org/10.1186/s13148-018-0450-y
Holliday H, Baker LA, Junankar SR et al (2018) Epigenomics of mammary gland development. Breast Cancer Res 20:100. https://doi.org/10.1186/s13058-018-1031-x
Hsieh CC, Trichopoulos D, Katsouyanni K, Yuasa S (1990) Age at menarche, age at menopause, height and obesity as risk factors for breast cancer: associations and interactions in an international case-control study. Int J Cancer 46:796–800. https://doi.org/10.1002/ijc.2910460508
Hu MC-T, Lee D-F, Xia W et al (2004) IκB Kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117:225–237. https://doi.org/10.1016/S0092-8674(04)00302-2
Hu Y-G, Hirasawa R, Hu J-L et al (2008) Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum Mol Genet 17:2654–2664. https://doi.org/10.1093/hmg/ddn165
Huang L, Hu B, Ni J et al (2016) Transcriptional repression of SOCS3 mediated by IL-6/STAT3 signaling via DNMT1 promotes pancreatic cancer growth and metastasis. J Exp Clin Cancer Res 35:27. https://doi.org/10.1186/s13046-016-0301-7
Huschtscha LI, Neumann AA, Noble JR, Reddel RR (2001) Effects of simian virus 40 T-antigens on normal human mammary epithelial cells reveal evidence for spontaneous alterations in addition to loss of p16(INK4a) expression. Exp Cell Res 265:125–134. https://doi.org/10.1006/excr.2001.5178
Iida T, Suetake I, Tajima S et al (2002) PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 7:997–1007. https://doi.org/10.1046/j.1365-2443.2002.00584.x
Intabli H, Flint MS, Qattan A et al (2019) 44P—the effect of cortisol on methylation patterns in breast cancer cell lines. Ann Oncol 30:v12–v13. https://doi.org/10.1093/annonc/mdz238.042
Irizarry RA, Ladd-Acosta C, Wen B et al (2009) Genome-wide methylation analysis of human colon cancer reveals similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41:178–186. https://doi.org/10.1038/ng.298
Ishida T, Yokoe T, Kasumi F et al (1992) Clinicopathologic characteristics and prognosis of breast cancer patients associated with pregnancy and lactation: analysis of case-control study in Japan. Jpn J Cancer Res 83:1143–1149. https://doi.org/10.1111/j.1349-7006.1992.tb02737.x
Izbicka E, Davidson KK, Lawrence RA et al (1999) 5,6-Dihydro-5’-azacytidine (DHAC) affects estrogen sensitivity in estrogen-refractory human breast carcinoma cell lines. Anticancer Res 19:1293–1298
Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140:1285–1295. https://doi.org/10.1083/jcb.140.6.1285
Jameera Begam A, Jubie S, Nanjan MJ (2017) Estrogen receptor agonists/antagonists in breast cancer therapy: a critical review. Bioorg Chem 71:257–274. https://doi.org/10.1016/j.bioorg.2017.02.011
Jia D, Jurkowska RZ, Zhang X et al (2007) Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248–251. https://doi.org/10.1038/nature06146
Jiang Y, Huang Y, Cheng C, Lu W, Zhang Y, Liu X, Zou L, Ben Q, Shen A (2011) Combination of thiazolidinedione and hydralazine suppresses proliferation and induces apoptosis by PPARγ up-expression in MDA-MB-231 cells. Exp Mol Pathol 91(3):768–774
Jin W, Chen L, Chen Y et al (2010) UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat 123:359–373. https://doi.org/10.1007/s10549-009-0652-2
Jinawath A, Miyake S, Yanagisawa Y et al (2005) Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem J 385:557–564. https://doi.org/10.1042/BJ20040684
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428. https://doi.org/10.1038/nrg816
Joo JE, Dowty JG, Milne RL et al (2018) Heritable DNA methylation marks associated with susceptibility to breast cancer. Nat Commun 9:1–12. https://doi.org/10.1038/s41467-018-03058-6
Junien C (2006) Impact of diets and nutrients/drugs on early epigenetic programming. J Inherit Metab Dis 29:359–365. https://doi.org/10.1007/s10545-006-0299-7
Kaaks R, Rinaldi S, Key TJ et al (2005) Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer 12:1071–1082. https://doi.org/10.1677/erc.1.01038
Kaminskas E, Farrell A, Abraham S et al (2005a) Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11:3604–3608. https://doi.org/10.1158/1078-0432.CCR-04-2135
Kaminskas E, Farrell AT, Wang Y-C et al (2005b) FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 10:176–182. https://doi.org/10.1634/theoncologist.10-3-176
Kandenwein JA, Park-Simon T-W, Schramm J, Simon M (2011) uPA/PAI-1 expression and uPA promoter methylation in meningiomas. J Neurooncol 103:533–539. https://doi.org/10.1007/s11060-010-0411-6
Kang ES, Park CW, Chung JH (2001a) Dnmt3b, de Novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem Biophys Res Commun 289:862–868. https://doi.org/10.1006/bbrc.2001.6057
Kang JH, Kim SJ, Noh D-Y et al (2001b) Methylation in the p53 promoter is a supplementary route to breast carcinogenesis: correlation between CpG methylation in the p53 promoter and the mutation of the p53 gene in the progression from ductal carcinoma in situ to invasive ductal carcinoma. Lab Invest 81:573–579. https://doi.org/10.1038/labinvest.3780266
Kang HJ, Yi YW, Hou S-J, et al (2015) Disruption of STAT3-DNMT1 interaction by SH-I-14 induces re-expression of tumor suppressor genes and inhibits growth of triple-negative breast tumor. Oncotarget 8:83457–83468. https://doi.org/https://doi.org/10.18632/oncotarget.4054
Kar S, Deb M, Sengupta D et al (2012) An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics 7:994–1007. https://doi.org/10.4161/epi.21568
Kareta MS, Botello ZM, Ennis JJ et al (2006) Reconstitution and mechanism of the stimulation of de Novo methylation by human DNMT3L. J Biol Chem 281:25893–25902. https://doi.org/10.1074/jbc.M603140200
Kim G-D, Ni J, Kelesoglu N et al (2002) Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J 21:4183–4195. https://doi.org/10.1093/emboj/cdf401
Kimura H, Shiota K (2003) Methyl-CpG-binding Protein, MeCP2, Is a target molecule for maintenance DNA methyltransferase, DNMT1. J Biol Chem 278:4806–4812. https://doi.org/10.1074/jbc.M209923200
Kishikawa S, Murata T, Kimura H et al (2002) Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem 269:2961–2970. https://doi.org/10.1046/j.1432-1033.2002.02972.x
Kiskinis E, Hallberg M, Christian M et al (2007) RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J 26:4831–4840. https://doi.org/10.1038/sj.emboj.7601908
Kong WY, Yee ZY, Mai CW, Fang CM, Abdullah S, Ngai SC (2019) Zebularine and trichostatin A sensitized human breast adenocarcinoma cells towards tumor necrosis factor-related apoptosis inducing ligand (TRAIL)-induced apoptosis. Heliyon 5(9):e02468
Kovalchuk O, Tryndyak VP, Montgomery B et al (2007) Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications, and aberrant microRNA expression. Cell Cycle 6:2010–2018. https://doi.org/10.4161/cc.6.16.4549
Krishna S, Shukla S, Lakra AD, Meeran SM, Siddiqi MI (2017) Identification of potent inhibitors of DNA methyltransferase 1 (DNMT1) through a pharmacophore-based virtual screening approach. J Mol Graph Model 75:174–188
Kruiswijk F, Labuschagne CF, Vousden KH (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16:393–405. https://doi.org/10.1038/nrm4007
Kuramochi-Miyagawa S, Watanabe T, Gotoh K et al (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22:908–917. https://doi.org/10.1101/gad.1640708
Kuwano Y, Kim HH, Abdelmohsen K et al (2008) MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol 28:4562–4575. https://doi.org/10.1128/MCB.00165-08
Laird PW (2003) The power and the promise of DNA methylation markers. Nat Rev Cancer 3:253–266. https://doi.org/10.1038/nrc1045
Lal A, Mazan-Mamczarz K, Kawai T et al (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23:3092–3102. https://doi.org/10.1038/sj.emboj.7600305
Landis-Piwowar KR, Milacic V, Dou QP (2008) Relationship between the methylation status of dietary flavonoids and their growth-inhibitory and apoptosis-inducing activities in human cancer cells. J Cell Biochem 105:514–523. https://doi.org/10.1002/jcb.21853
Laptenko O, Prives C (2017) p53: master of life, death, and the epigenome. Genes Dev 31:955–956. https://doi.org/10.1101/gad.302364.117
Lavoie G, Estève P-O, Laulan NB et al (2011) PKC isoforms interact with and phosphorylate DNMT1. BMC Biol 9:31. https://doi.org/10.1186/1741-7007-9-31
Lavoie G, St-Pierre Y (2011) Phosphorylation of human DNMT1: implication of cyclin-dependent kinases. Biochem Biophys Res Commun 409:187–192. https://doi.org/10.1016/j.bbrc.2011.04.115
Le X-F, Spizzo R, Mao M et al (2010) Abstract 2051: DNA (cytosine-5-)-methyltransferases 3A (DNMT3A) is a direct target of miR-194 in breast cancer. Cancer Res 70:2051–2051. https://doi.org/10.1158/1538-7445.AM10-2051
Lee B, Muller MT (2009) SUMOylation enhances DNA methyltransferase 1 activity. Biochem J 421:449–461. https://doi.org/10.1042/BJ20090142
Lee H, Zhang P, Herrmann A et al (2012) Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. PNAS 109:7765–7769. https://doi.org/10.1073/pnas.1205132109
Leick MB, Shoff CJ, Wang EC et al (2011) Loss of imprinting of IGF2 and the epigenetic progenitor model of cancer. Am J Stem Cells 1:59–74
Leng F, Yu J, Zhang C et al (2018) Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4 DCAF5 ubiquitin ligase. Nat Commun 9:1–17. https://doi.org/10.1038/s41467-018-04019-9
Levy D (2019) Lysine methylation signaling of non-histone proteins in the nucleus. Cell Mol Life Sci 76:2873–2883. https://doi.org/10.1007/s00018-019-03142-0
Levy NS, Chung S, Furneaux H, Levy AP (1998) Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem 273:6417–6423. https://doi.org/10.1074/jbc.273.11.6417
Li Y, Liu L, Andrews LG, Tollefsbol TO (2009) Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer 125(2):286–296
Li L, Lee K-M, Han W et al (2010a) Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet 19:4273–4277. https://doi.org/10.1093/hmg/ddq351
Li Y, Yuan Y-Y, Meeran SM, Tollefsbol TO (2010b) Synergistic epigenetic reactivation of estrogen receptor-alpha (ERalpha) by combined green tea polyphenol and histone deacetylase inhibitor in ERalpha-negative breast cancer cells. Mol Cancer 9:274. https://doi.org/10.1186/1476-4598-9-274
Li Y, Chen H, Hardy TM, Tollefsbol TO (2013a) Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS ONE. https://doi.org/10.1371/journal.pone.0054369
Li Z, Du L, Li C, Wu W (2013b) Human chorionic gonadotropin β induces cell motility via ERK1/2 and MMP-2 activation in human glioblastoma U87MG cells. J Neurooncol 111:237–244. https://doi.org/10.1007/s11060-012-1017-y
Li Z, Li C, Du L et al (2013c) Human chorionic gonadotropin β induces migration and invasion via activating ERK1/2 and MMP-2 in human prostate cancer DU145 Cells. PLoS ONE. https://doi.org/10.1371/journal.pone.0054592
Li X, Zha Q, Ren Z et al (2015) Mechanisms of breast cancer resistance to anthracyclines or taxanes: an overview of the proposed roles of noncoding RNA. Curr Opin Oncol 27:457–465. https://doi.org/10.1097/CCO.0000000000000235
Li Y, Tollefsbol TO (2010) Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem 17:2141–2151. https://doi.org/10.2174/092986710791299966
Liao X-H, Lu D-L, Wang N et al (2014) Estrogen receptor α mediates proliferation of breast cancer MCF–7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J 281:927–942. https://doi.org/10.1111/febs.12658
Lin R-K, Hsu H-S, Chang J-W et al (2007) Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer 55:205–213. https://doi.org/10.1016/j.lungcan.2006.10.022
Lin R-K, Hsieh Y-S, Lin P et al (2010a) The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest 120:521–532. https://doi.org/10.1172/JCI40706
Lin R-K, Wu C-Y, Chang J-W et al (2010b) Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res 70:5807–5817. https://doi.org/10.1158/0008-5472.CAN-09-4161
Ling Y, Sankpal UT, Robertson AK et al (2004) Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res 32:598–610. https://doi.org/10.1093/nar/gkh195
Lippert C, Seeger H, Mueck AO (2003) The effect of endogenous estradiol metabolites on the proliferation of human breast cancer cells. Life Sci 72:877–883. https://doi.org/10.1016/S0024-3205(02)02305-6
Liu S, Shen T, Huynh L et al (2005) Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res 65:1277–1284. https://doi.org/10.1158/0008-5472.CAN-04-4532
Liu X, Li C, Zhang R et al (2018) The EZH2- H3K27me3-DNMT1 complex orchestrates epigenetic silencing of the wwc1 gene, a Hippo/YAP pathway upstream effector, in breast cancer epithelial cells. Cell Signal 51:243–256. https://doi.org/10.1016/j.cellsig.2018.08.011
López de Silanes I, Gorospe M, Taniguchi H et al (2009) The RNA-binding protein HuR regulates DNA methylation through stabilization of DNMT3b mRNA. Nucleic Acids Res 37:2658–2671. https://doi.org/10.1093/nar/gkp123
Lorente A, Mueller W, Urdangarín E et al (2009) RASSF1A, BLU, NORE1A, PTEN and MGMT expression and promoter methylation in gliomas and glioma cell lines and evidence of deregulated expression of de novo DNMTs. Brain Pathol 19:279–292. https://doi.org/10.1111/j.1750-3639.2008.00185.x
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838. https://doi.org/10.1038/nature03702
Lukas J, Gao D-Q, Keshmeshian M et al (2001) Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res 61:3212–3219
Luo RX, Postigo AA, Dean DC (1998) Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473. https://doi.org/10.1016/S0092-8674(00)80940-X
Macaluso M, Cinti C, Russo G et al (2003) pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor- α in breast cancer. Oncogene 22:3511–3517. https://doi.org/10.1038/sj.onc.1206578
Macaluso M, Montanari M, Noto PB et al (2007) Epigenetic modulation of estrogen receptor-α by pRb family proteins: a novel mechanism in breast cancer. Cancer Res 67:7731–7737. https://doi.org/10.1158/0008-5472.CAN-07-1476
Macias H, Hinck L (2012) Mammary gland development. WIREs. Dev Biol 1:533–557. https://doi.org/10.1002/wdev.35
MacLeod AR, Rouleau J, Szyf M (1995) Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem 270:11327–11337. https://doi.org/10.1074/jbc.270.19.11327
Makino T, Yamasaki M, Miyata H et al (2010) P53 mutation status predicts pathological response to chemoradiotherapy in locally advanced esophageal cancer. Ann Surg Oncol 17:804–811. https://doi.org/10.1245/s10434-009-0786-9
Maleva Kostovska I, Jakimovska M, Popovska-Jankovic K et al (2018) TIMP3 promoter methylation represents an epigenetic marker of BRCA1ness breast cancer tumours. Pathol Oncol Res 24:937–940. https://doi.org/10.1007/s12253-018-0398-4
Margot JB, Cardoso MC, Leonhardt H (2001) Mammalian DNA methyltransferases show different subnuclear distributions. J Cell Biochem 83:373–379. https://doi.org/10.1002/jcb.1236
Maruti SS, Ulrich CM, White E (2009) Folate and one-carbon metabolism nutrients from supplements and diet in relation to breast cancer risk. Am J Clin Nutr 89:624–633. https://doi.org/10.3945/ajcn.2008.26568
McCabe MT, Davis JN, Day ML (2005) Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res 65:3624–3632. https://doi.org/10.1158/0008-5472.CAN-04-2158
Mei Y, Clark D, Mao L (2013) Novel dimensions of piRNAs in cancer. Cancer Lett 336:46–52. https://doi.org/10.1016/j.canlet.2013.04.008
Melhem A, Yamada SD, Fleming GF et al (2009) Administration of glucocorticoids to ovarian cancer patients is associated with expression of the anti-apoptotic genes SGK1 and MKP1/DUSP1 in ovarian tissues. Clin Cancer Res 15:3196–3204. https://doi.org/10.1158/1078-0432.CCR-08-2131
Melo SA, Esteller M (2011) Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett 585:2087–2099. https://doi.org/10.1016/j.febslet.2010.08.009
Michael D, Oren M (2003) The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol 13:49–58. https://doi.org/10.1016/S1044-579X(02)00099-8
Migliaccio A, Piccolo D, Castoria G et al (1998) Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. The EMBO Journal 17:2008–2018. https://doi.org/10.1093/emboj/17.7.2008
Mikosz CA, Brickley DR, Sharkey MS et al (2001) Glucocorticoid receptor-mediated protection from apoptosis is associated with induction of the serine/threonine survival kinase gene, SGK-1. J Biol Chem 276:16649–16654. https://doi.org/10.1074/jbc.M010842200
Miller LD, Smeds J, George J et al (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. PNAS 102:13550–13555
Ming X-F, Stoecklin G, Lu M et al (2001) Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol 21:5778–5789. https://doi.org/10.1128/MCB.21.17.5778-5789.2001
Mirza S, Sharma G, Pandya P, Ralhan R (2010) Demethylating agent 5-aza-2-deoxycytidine enhances susceptibility of breast cancer cells to anticancer agents. Mol Cell Biochem 342(1–2):101–109
Mizuno S, Chijiwa T, Okamura T et al (2001) Expression of DNA methyltransferases DNMT1,3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97:1172–1179. https://doi.org/10.1182/blood.V97.5.1172
Munro AJ, Lain S, Lane DP (2005) P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer 92:434–444. https://doi.org/10.1038/sj.bjc.6602358
Muromoto R, Sugiyama K, Takachi A et al (2004) Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J Immunol 172:2985–2993. https://doi.org/10.4049/jimmunol.172.5.2985
Muvarak NE, Chowdhury K, Xia L et al (2016) Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents—a potential therapy for cancer. Cancer Cell 30:637–650. https://doi.org/10.1016/j.ccell.2016.09.002
Myant K, Stancheva I (2008) LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol 28:215–226. https://doi.org/10.1128/MCB.01073-07
Newman EM, Morgan RJ, Kummar S, Beumer JH, Blanchard MS, Ruel C, El-Khoueiry AB, Carroll MI, Hou JM, Li C, Lenz HJ, Eiseman Doroshow JH (2015) A phase I, pharmacokinetic, and pharmacodynamic evaluation of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine, administered with tetrahydrouridine. Cancer Chemother Pharmacol 75(3):537–546
Ng EKO, Tsang WP, Ng SSM et al (2009) MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 101:699–706. https://doi.org/10.1038/sj.bjc.6605195
Ng EKO, Li R, Shin VY et al (2014) MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumor Biol 35:2591–2598. https://doi.org/10.1007/s13277-013-1341-7
Niculescu MD, Lupu DS (2011) Nutritional influence on epigenetics and effects on longevity. Curr Opin Clin Nutr Metab Care 14:35–40. https://doi.org/10.1097/MCO.0b013e328340ff7c
Nielsen NR, Stahlberg C, Strandberg-Larsen K et al (2008) Are work-related stressors associated with diagnosis of more advanced stages of incident breast cancers? Cancer Causes Control 19:297–303. https://doi.org/10.1007/s10552-007-9092-7
Nishiyama A, Mulholland CB, Bultmann S et al (2020) Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat Commun 11:1–17. https://doi.org/10.1038/s41467-020-15006-4
Obradović MMS, Hamelin B, Manevski N et al (2019) Glucocorticoids promote breast cancer metastasis. Nature 567:540–544. https://doi.org/10.1038/s41586-019-1019-4
Okada M, Lee L, Maekawa R et al (2016) Epigenetic changes of the CYP11A1 promoter region in granulosa cells undergoing luteinization during ovulation in female rats. Endocrinology 157:3344–3354. https://doi.org/10.1210/en.2016-1264
Okano M, Bell DW, Haber DA, Li E (1999) Dna methyltransferases DNMT3A AND DNMT3B are essential for de novo methylation and mammalian development. Cell 99:247–257. https://doi.org/10.1016/S0092-8674(00)81656-6
Okumura N, Saji S, Eguchi H et al (2002) Distinct promoter usage of mdm2 gene in human breast cancer. Oncol Rep 9:557–563. https://doi.org/10.3892/or.9.3.557
Olivier M, Langer A, Carrieri P et al (2006) The clinical value of somatic TP53 gene mutations in 1794 patients with breast cancer. Clin Cancer Res 12:1157–1167. https://doi.org/10.1158/1078-0432.CCR-05-1029
Pakneshan P, Szyf M, Rabbani SA (2005) Hypomethylation of urokinase (uPA) promoter in breast and prostate cancer: Prognostic and therapeutic implications. https://www.ingentaconnect.com/content/ben/ccdt/2005/00000005/00000007/art00002. Accessed 31 Mar 2020
Palakurthy RK, Wajapeyee N, Santra MK et al (2009) Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell 36:219–230. https://doi.org/10.1016/j.molcel.2009.10.009
Patra SK (2008) Ras regulation of DNA-methylation and cancer. Exp Cell Res 314:1193–1201. https://doi.org/10.1016/j.yexcr.2008.01.012
Pauwels S, Duca RC, Devlieger R et al (2016) Maternal methyl-group donor Intake and global DNA (Hydroxy)methylation before and during Pregnancy. Nutrients 8:474. https://doi.org/10.3390/nu8080474
Peifer M, Fernández-Cuesta L, Sos ML et al (2012) Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44:1104–1110. https://doi.org/10.1038/ng.2396
Peng L, Yuan Z, Ling H et al (2011) SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol 31:4720–4734. https://doi.org/10.1128/MCB.06147-11
Peng W-X, Koirala P, Zhang W et al (2020) lncRNA RMST enhances DNMT3 expression through interaction with HuR. Mol Ther 28:9–18. https://doi.org/10.1016/j.ymthe.2019.09.024
Peterson EJ, Bögler O, Taylor SM (2003) P53-mediated repression of DNA methyltransferase 1 expression by specific dna binding. Cancer Res 63:6579–6582
Petitjean A, Mathe E, Kato S et al (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28:622–629. https://doi.org/10.1002/humu.20495
Pichler G, Wolf P, Schmidt CS et al (2011) Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J Cell Biochem 112:2585–2593. https://doi.org/10.1002/jcb.23185
Planeix F, Siraj M-A, Bidard F-C et al (2015) Endothelial follicle-stimulating hormone receptor expression in invasive breast cancer and vascular remodeling at tumor periphery. J Exp Clin Cancer Res 34:12. https://doi.org/10.1186/s13046-015-0128-7
Pradhan N, Parbin S, Kar S et al (2019) Epigenetic silencing of genes enhanced by collective role of reactive oxygen species and MAPK signaling downstream ERK/Snail axis: Ectopic application of hydrogen peroxide repress CDH1 gene by enhanced DNA methyltransferase activity in human breast cancer. Biochim Biophys Acta BBA Mol Basis Dis 1865:1651–1665. https://doi.org/10.1016/j.bbadis.2019.04.002
Pradhan S, Bacolla A, Wells RD, Roberts RJ (1999) Recombinant human DNA (Cytosine-5) methyltransferase I. expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 274:33002–33010. https://doi.org/10.1074/jbc.274.46.33002
Pulliam N, Fang F, Ozes AR, Tang J, Adewuyi A, Keer H, Lyons J, Baylin SB, Matei D, Nakshatri H, Rassool FV, Miller KD, Nephew KP (2018) An Effective Epigenetic-PARP Inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA Mutations. Clin Cancer Res 24(13):3163–3175
Qian C, Li S, Jakoncic J et al (2008) Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J Biol Chem 283:34490–34494. https://doi.org/10.1074/jbc.C800169200
Quintás-Cardama A, Kantarjian H, Estrov Z et al (2012) Therapy with the histone deacetylase inhibitor pracinostat for patients with myelofibrosis. Leuk Res 36:1124–1127. https://doi.org/10.1016/j.leukres.2012.03.003
Radpour R, Kohler C, Haghighi MM et al (2009) Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene 28:2969–2978. https://doi.org/10.1038/onc.2009.149
Ramsahoye BH, Biniszkiewicz D, Lyko F et al (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. PNAS 97:5237–5242. https://doi.org/10.1073/pnas.97.10.5237
Rauscher GH, Kresovich JK, Poulin M et al (2015) Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer 15:816. https://doi.org/10.1186/s12885-015-1777-9
Reinders J, Sickmann A (2007) Modificomics: posttranslational modifications beyond protein phosphorylation and glycosylation. Biomol Eng 24:169–177. https://doi.org/10.1016/j.bioeng.2007.03.002
Ren S, Rollins BJ (2004) Cyclin C/Cdk3 promotes Rb-dependent G0 exit. Cell 117:239–251. https://doi.org/10.1016/S0092-8674(04)00300-9
Rhee I, Bachman KE, Park BH et al (2002) DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552–556. https://doi.org/10.1038/416552a
Rice LW (2010) Hormone prevention strategies for breast, endometrial and ovarian cancers. Gynecol Oncol 118:202–207. https://doi.org/10.1016/j.ygyno.2010.03.014
Richard J, Sainsbury C, GeoffreyK N et al (1987) Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. The Lancet 329:1398–1402. https://doi.org/10.1016/S0140-6736(87)90593-9
Richter A, Roolf C, Hamed M et al (2019) Combined Casein Kinase II inhibition and epigenetic modulation in acute B-lymphoblastic leukemia. BMC Cancer 19:202. https://doi.org/10.1186/s12885-019-5411-0
Rijnkels M, Kabotyanski E, Montazer-Torbati MB et al (2010) The epigenetic landscape of mammary gland development and functional differentiation. J Mammary Gland Biol Neoplasia 15:85–100. https://doi.org/10.1007/s10911-010-9170-4
Robertson JFR, Blamey RW (2003) The use of gonadotrophin-releasing hormone (GnRH) agonists in early and advanced breast cancer in pre- and perimenopausal women. Eur J Cancer 39:861–869. https://doi.org/10.1016/S0959-8049(02)00810-9
Robertson KD, Ait-Si-Ali S, Yokochi T et al (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 25:338–342. https://doi.org/10.1038/77124
Rock CL, Demark-Wahnefried W (2002) Nutrition and survival after the diagnosis of breast cancer : A review of the evidence. J Clin Oncol 20:3302–3316. https://doi.org/10.1200/JCO.2002.03.008
Roll JD, Rivenbark AG, Jones WD, Coleman WB (2008) DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer 7:15. https://doi.org/10.1186/1476-4598-7-15
Rondelet G, Fleury L, Faux C et al (2017) Inhibition studies of DNA methyltransferases by maleimide derivatives of RG108 as non-nucleoside inhibitors. Fut Med Chem 9:1465–1481. https://doi.org/10.4155/fmc-2017-0074
Rotili D, Tarantino D, Marrocco B et al (2014) Properly substituted analogues of BIX-01294 Lose inhibition of G9a histone methyltransferase and gain selective anti-NA methyltransferase 3A activity. PLoS ONE. https://doi.org/10.1371/journal.pone.0096941
Russo IH, Russo J (2011) Pregnancy-induced changes in breast cancer risk. J Mammary Gland Biol Neoplasia 16:221. https://doi.org/10.1007/s10911-011-9228-y
Russo IH, Russo J (2007) Primary prevention of breast cancer by hormone-induced differentiation. In: Senn H-J, Kapp U (eds) Cancer prevention. Springer, Berlin, Heidelberg, pp 111–130
Sacco F, Humphrey SJ, Cox J et al (2016) Glucose-regulated and drug-perturbed phosphoproteome reveals molecular mechanisms controlling insulin secretion. Nat Commun 7:1–13. https://doi.org/10.1038/ncomms13250
Sadikovic B, Al-Romaih K, Squire JA, Zielenska M (2008) Cause and consequences of genetic and epigenetic alterations in human cancer. https://www.ingentaconnect.com/content/ben/cg/2008/00000009/00000006/art00004. Accessed 31 Mar 2020
San José-Enériz E, Agirre X, Rabal O et al (2017) Discovery of first-in-class reversible dual small molecule inhibitors against G9a and DNMTs in hematological malignancies. Nat Commun 8:1–10. https://doi.org/10.1038/ncomms15424
Sanchez AM, Flamini MI, Russo E et al (2016) LH and FSH promote migration and invasion properties of a breast cancer cell line through regulatory actions on the actin cytoskeleton. Mol Cell Endocrinol 437:22–34. https://doi.org/10.1016/j.mce.2016.08.009
Sandhu R, Rivenbark AG, Coleman WB (2012) Loss of post-transcriptional regulation of DNMT3b by microRNAs: A possible molecular mechanism for the hypermethylation defect observed in a subset of breast cancer cell lines. Int J Oncol 41:721–732. https://doi.org/10.3892/ijo.2012.1505
Santen RJ, Boyd NF, Chlebowski RT et al (2007) Critical assessment of new risk factors for breast cancer: considerations for development of an improved risk prediction model. Endocr Relat Cancer 14:169–187. https://doi.org/10.1677/ERC-06-0045
Santen RJ, Allred DC, Ardoin SP et al (2010) Postmenopausal hormone therapy: an endocrine society scientific statement. J Clin Endocrinol Metab 95:s1–s66. https://doi.org/10.1210/jc.2009-2509
Schmitt CA, Fridman JS, Yang M et al (2002) A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109:335–346. https://doi.org/10.1016/S0092-8674(02)00734-1
Sdek P, Ying H, Chang DLF et al (2005) MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell 20:699–708. https://doi.org/10.1016/j.molcel.2005.10.017
Seenappa V, Das B, Joshi MB, Satyamoorthy K (2016) Context dependent regulation of human phosphoenolpyruvate carboxykinase isoforms by dna promoter methylation and rna stability: D ISTINCT M ECHANISMS C ONTROL PEPCK I SOFORMS. J Cell Biochem 117:2506–2520. https://doi.org/10.1002/jcb.25543
Segura-Pacheco B, Trejo-Becerril C, Perez-Cardenas E et al (2003) Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin Cancer Res 9:1596–160
Sengupta S, Jang B-C, Wu M-T et al (2003) The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J Biol Chem 278:25227–25233. https://doi.org/10.1074/jbc.M301813200
Seo J, Li L, Small D (2019) Dissociation of the DNMT3A-HDAC1 repressor complex induces PD-L1 expression. Blood 134:3759–3759. https://doi.org/10.1182/blood-2019-130339
Sephton SE, Sapolsky RM, Kraemer HC, Spiegel D (2000) Diurnal cortisol rhythm as a predictor of breast cancer survival. J Natl Cancer Inst 92:994–1000. https://doi.org/10.1093/jnci/92.12.994
Severin J, Lizio M, Harshbarger J et al (2014) Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol 32:217–219. https://doi.org/10.1038/nbt.2840
Shamay M, Greenway M, Liao G et al (2010) De Novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J Biol Chem 285:36377–36386. https://doi.org/10.1074/jbc.M110.155721
Shao Z, Robbins PD (1995) Differential regulation of E2F and Sp1-mediated transcription by G1 cyclins. Oncogene 10:221–228
Sharma M, Chuang WW, Sun Z (2002) Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through GSK3β inhibition and nuclear β-catenin accumulation. J Biol Chem 277:30935–30941. https://doi.org/10.1074/jbc.M201919200
Shen Y, Katsaros D, Loo LWM et al (2015) Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget 6:8579–8592. https://doi.org/10.18632/oncotarget.3287
Shi Y, Li J, Liu Y et al (2015) The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer 14:51. https://doi.org/10.1186/s12943-015-0318-0
Si X, Liu Y, Lv J et al (2016) ERα propelled aberrant global DNA hypermethylation by activating the DNMT1 gene to enhance anticancer drug resistance in human breast cancer cells. Oncotarget 7:20966–20980. https://doi.org/10.18632/oncotarget.8038
Siedlecki P, Boy RG, Musch T et al (2006) Discovery of two novel, small-molecule inhibitors of DNA methylation. J Med Chem 49:678–683. https://doi.org/10.1021/jm050844z
Singh BN, Singh HB, Singh A et al (2014) Dietary phytochemicals alter epigenetic events and signaling pathways for inhibition of metastasis cascade: phytoblockers of metastasis cascade. Cancer Metastasis Rev 33:41–85. https://doi.org/10.1007/s10555-013-9457-1
Sinha D, Biswas J, Sung B, Aggarwal BB, Bishayee A (2012) Chemopreventive and chemotherapeutic potential of curcumin in breast cancer. Curr Drug Targets 13:1799–1819. https://doi.org/10.2174/138945012804545632
Skor MN, Wonder EL, Kocherginsky M et al (2013) Glucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancer. Clin Cancer Res 19:6163–6172. https://doi.org/10.1158/1078-0432.CCR-12-3826
Smit L, Berns K, Spence K et al (2015) An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget 7:2596–2610. https://doi.org/10.18632/oncotarget.6354
Smith LH, Dalrymple JL, Leiserowitz GS et al (2001) Obstetrical deliveries associated with maternal malignancy in California, 1992 through 1997. Am J Obstet Gynecol 184:1504–1513. https://doi.org/10.1067/mob.2001.114867
Song I-S, Tatebe S, Dai W, Kuo MT (2005) Delayed mechanism for induction of γ-glutamylcysteine synthetase heavy subunit mRNA stability by oxidative stress involving p38 mitogen-activated protein kinase signaling. J Biol Chem 280:28230–28240. https://doi.org/10.1074/jbc.M413103200
Steeg PS, Ouatas T, Halverson D et al (2003) Metastasis suppressor genes: Basic biology and potential clinical use. Clinical Breast Cancer 4:51–62. https://doi.org/10.3816/CBC.2003.n.012
Ströfer M, Jelkmann W, Depping R (2011) Curcumin decreases survival of Hep3B liver and MCF-7 breast cancer cells: The role of HIF. Strahlenther Onkol 187:393–400. https://doi.org/10.1007/s00066-011-2248-0
Suetake I, Shinozaki F, Miyagawa J et al (2004) DNMT3L stimulates the DNA methylation activity of DNMT3A and DNMT3B through a direct interaction. J Biol Chem 279:27816–27823. https://doi.org/10.1074/jbc.M400181200
Sugiyama Y, Hatano N, Sueyoshi N et al (2010) The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1δ/ε. Biochem J 427:489–497. https://doi.org/10.1042/BJ20091856
Sunters A, Madureira PA, Pomeranz KM et al (2006) Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res 66:212–220. https://doi.org/10.1158/0008-5472.CAN-05-1997
Taketo MM (2004) Shutting down Wnt signal–activated cancer. Nat Genet 36:320–322. https://doi.org/10.1038/ng0404-320
Tan J, Gu Y, Zhang X et al (2013) Hypermethylation of CpG islands is more prevalent than hypomethylation across the entire genome in breast carcinogenesis. Clin Exp Med 13:1–9. https://doi.org/10.1007/s10238-011-0173-2
Tang Y-A, Lin R-K, Tsai Y-T et al (2012) MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res 18:4325–4333. https://doi.org/10.1158/1078-0432.CCR-11-2617
Tartakover-Matalon S, Mizrahi A, Epstein G et al (2010) Breast cancer characteristics are modified by first trimester human placenta: in vitro co-culture study. Hum Reprod 25:2441–2454. https://doi.org/10.1093/humrep/deq227
Tatematsu K, Yamazaki T, Ishikawa F (2000) MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677–688. https://doi.org/10.1046/j.1365-2443.2000.00359.x
Tempera I, Lieberman PM (2014) Epigenetic regulation of EBV persistence and oncogenesis. Semin Cancer Biol 26:22–29. https://doi.org/10.1016/j.semcancer.2014.01.003
Teschendorff AE, Gao Y, Jones A et al (2016) DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat Commun 7:1–12. https://doi.org/10.1038/ncomms10478
Teulings FA, van Gilse HA (1977) Demonstration of glucocorticoid receptors in human mammary carcinomas. Horm Res 8:107–116. https://doi.org/10.1159/000178787
Thanos J, Cotterchio M, Boucher BA et al (2006) Adolescent dietary phytoestrogen intake and breast cancer risk (Canada). Cancer Causes Control 17:1253–1261. https://doi.org/10.1007/s10552-006-0062-2
Thomas NSB (2012) The STAT3-DNMT1 connection. JAK-STAT 1:257–260. https://doi.org/10.4161/jkst.22436
Tran H, Maurer F, Nagamine Y (2003) Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol 23:7177–7188. https://doi.org/10.1128/MCB.23.20.7177-7188.2003
Tsai C-N, Tsai C-L, Tse K-P et al (2002) The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. PNAS 99:10084–10089. https://doi.org/10.1073/pnas.152059399
Tuo Y-L, Li X-M, Luo J (2015) Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci 19:3403–3411
Uysal F, Ozturk S, Akkoyunlu G (2018) Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J Assist Reprod Genet 35:503–513. https://doi.org/10.1007/s10815-017-1087-z
Verde G, De Llobet LI, Wright RHG et al (2018) Unliganded progesterone receptor governs estrogen receptor gene expression by regulating DNA methylation in breast cancer cells. Cancers 10:371. https://doi.org/10.3390/cancers10100371
Verger A, Perdomo J, Crossley M (2003) Modification with SUMO. A role in transcriptional regulation. EMBO Rep 4:137–142. https://doi.org/10.1038/sj.embor.embor738
Wang J, Hevi S, Kurash JK et al (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41:125–129. https://doi.org/10.1038/ng.268
Wang W, Caldwell MC, Lin S et al (2000a) HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 19:2340–2350. https://doi.org/10.1093/emboj/19.10.2340
Wang W, Furneaux H, Cheng H et al (2000b) HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20:760–769. https://doi.org/10.1128/MCB.20.3.760-769.2000
Wang Y, Zhou Y, Graves DT (2014) FOXO transcription factors: their clinical significance and regulation. Biomed Res Int. https://doi.org/10.1155/2014/925350
Wang YA, Kamarova Y, Shen KC et al (2005) DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol Ther 4:1138–1143. https://doi.org/10.4161/cbt.4.10.2073
Walters RS, Theriault RL, Holmes FA et al (1992) Phase II trial of fazarabine (ARA-AC, arabinosyl-5-azacytosine) in metastatic breast cancer. Invest New Drugs 10:43–44. https://doi.org/10.1007/BF01275480
Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81:323–330. https://doi.org/10.1016/0092-8674(95)90385-2
Wen Y-Y, Liu W-T, Sun H-R et al (2017) IGF-1-mediated PKM2/β-catenin/miR-152 regulatory circuit in breast cancer. Sci Rep 7:1–10. https://doi.org/10.1038/s41598-017-15607-y
Wu J, Shuang Z, Zhao J et al (2018) Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother 97:1275–1281. https://doi.org/10.1016/j.biopha.2017.11.055
Wu Q, Odwin-Dacosta S, Cao S et al (2019) Estrogen down regulates COMT transcription via promoter DNA methylation in human breast cancer cells. Toxicol Appl Pharmacol 367:12–22. https://doi.org/10.1016/j.taap.2019.01.016
Wu W, Chaudhuri S, Brickley DR et al (2004) Microarray analysis reveals glucocorticoid-regulated survival genes that are associated with inhibition of apoptosis in breast epithelial cells. Cancer Res 64:1757–1764. https://doi.org/10.1158/0008-5472.CAN-03-2546
Wu W, Walker AM (2006) Human chorionic gonadotropin β (HCGβ) down-regulates E-cadherin and promotes human prostate carcinoma cell migration and invasion. Cancer 106:68–78. https://doi.org/10.1002/cncr.21549
Xie Q, Bai Q, Zou L-Y et al (2014) Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosom Cancer 53:422–431. https://doi.org/10.1002/gcc.22154
Xie T, Yu J, Fu W et al (2019) Insight into the selective binding mechanism of DNMT1 and DNMT3A inhibitors: a molecular simulation study. Phys Chem Chem Phys 21:12931–12947. https://doi.org/10.1039/C9CP02024A
Xing X-B, Cai W-B, Luo L et al (2013) The prognostic value of p16 hypermethylation in cancer: a meta-analysis. PLoS ONE. https://doi.org/10.1371/journal.pone.0066587
Xiong Y, Dowdy SC, Xue A et al (2005) Opposite alterations of DNA methyltransferase gene expression in endometrioid and serous endometrial cancers. Gynecol Oncol 96:601–609. https://doi.org/10.1016/j.ygyno.2004.11.047
Xu M, Zhu J, Liu S et al (2019) FOXD3, frequently methylated in colorectal cancer, acts as a tumor suppressor and induces tumor cell apoptosis under ER stress via p53. Carcinogenesis. https://doi.org/10.1093/carcin/bgz198
Xue Y, Zhou F, Fu C et al (2006) SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res 34:W254–W257. https://doi.org/10.1093/nar/gkl207
Yamagata Y, Asada H, Tamura I et al (2009) DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum Reprod 24:1126–1132. https://doi.org/10.1093/humrep/dep015
Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE (2003) Specific Inhibition of DNMT1 by Antisense Oligonucleotides Induces Re-expression of Estrogen Receptor a (ER) in ER-negative Human Breast Cancer Cell Lines. Cancer Biol Ther 2(5):552–556
Yang Y-C, Tang Y-A, Shieh J-M et al (2014) DNMT3B overexpression by deregulation of FOXO3A-mediated transcription repression and MDM2 overexpression in lung cancer. J Thorac Oncol 9:1305–1315. https://doi.org/10.1097/JTO.0000000000000240
Yang Z, Guo F, Albers AE, Sehouli J, Kaufmann AM (2019) Disulfiram modulates ROS accumulation and overcomes synergistically cisplatin resistance in breast cancer cell lines. Biomed Pharmacother 113:108727
Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809. https://doi.org/10.1038/nrc2734
Yu J, Qin B, Moyer AM et al (2018) DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Investig 128:2376–2388. https://doi.org/10.1172/JCI97924
Yu Z, Xiao Q, Zhao L et al (2015) DNA methyltransferase 1/3a overexpression in sporadic breast cancer is associated with reduced expression of estrogen receptor-alpha/breast cancer susceptibility gene 1 and poor prognosis: CORRELATION OF DNMT1/3A WITH PROGNOSIS IN BREAST CANCER. Mol Carcinog 54:707–719. https://doi.org/10.1002/mc.22133
Zeisel SH (2009) Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr 89:1488S-1493S. https://doi.org/10.3945/ajcn.2009.27113B
Zhang H, Gao Q, Tan S et al (2019) SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1. Nucleic Acids Res 47:9053–9068. https://doi.org/10.1093/nar/gkz626
Zhang HS, Gavin M, Dahiya A et al (2000) Exit from G1 and SpPhase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79–89. https://doi.org/10.1016/S0092-8674(00)80625-X
Zhang L, Yang W, Zhu X, Wei C (2016) p53 inhibits the expression of p125 and the methylation of POLD1 gene promoter by downregulating the Sp1-induced DNMT1 activities in breast cancer. Onco Targets Ther 9:1351–1360. https://doi.org/10.2147/OTT.S98713
Zhang Q, Wang HY, Marzec M et al (2005) STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. PNAS 102:6948–6953. https://doi.org/10.1073/pnas.0501959102
Zhang X, Yee D (2000) Tyrosine kinase signalling in breast cancer: insulin-like growth factors and their receptors in breast cancer. Breast Cancer Res 2:170. https://doi.org/10.1186/bcr50
Zhou Q, Agoston AT, Atadja P et al (2008) Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 6:873–883. https://doi.org/10.1158/1541-7786.MCR-07-0330
Zhu X, Shan L, Wang F et al (2015) Hypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast Cancer Res Treat 150:479–486. https://doi.org/10.1007/s10549-015-3338-y
Zou Y, Tsai W-B, Cheng C-J et al (2008) Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res 10:R21. https://doi.org/10.1186/bcr1872
Acknowledgements
Authors thank TIFAC Core, Government of India and Manipal Academy for Higher Education, Manipal for infrastructure and 6242-P81/RGCB/PMD/DBT/MNBJ/2015 Cancer biology-Pilot studies grant from Department of Biotechnology, Government of India for funding. Authors thank Dr. K. Satyamoorthy for inputs and guidance. Mangala Hegde is financially supported by Dr. TMA Pai Fellowship, MAHE, Manipal and Senior Research Fellowship, CSIR, Govt of India.
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hegde, M., Joshi, M.B. Comprehensive analysis of regulation of DNA methyltransferase isoforms in human breast tumors. J Cancer Res Clin Oncol 147, 937–971 (2021). https://doi.org/10.1007/s00432-021-03519-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03519-4