Abstract
Key message
Glutamine synthetase TaGSr-4B is a candidate gene for a QTL of thousand grain weight on 4B, and the gene marker is ready for wheat breeding.
Abstract
A QTL for thousand grain weight (TGW) in wheat was previously mapped on chromosome 4B in a DH population of Westonia × Kauz. For identifying the candidate genes of the QTL, wheat 90 K SNP array was used to saturate the existing linkage map, and four field trials plus one glasshouse experiment over five locations were conducted to refine the QTL. Three nitrogen levels were applied to two of those field trials, resulting in a TGW phenotype data set from nine environments. A robust TGW QTL cluster including 773 genes was detected in six environments with the highest LOD value of 13.4. Based on differentiate gene expression within the QTL cluster in an RNAseq data of Westonia and Kauz during grain filling, a glutamine synthesis gene (GS: TaGSr-4B) was selected as a potential candidate gene for the QTL. A SNP on the promoter region between Westonia and Kauz was used to develop a cleaved amplified polymorphic marker for TaGSr-4B gene mapping and QTL reanalysing. As results, TGW QTL appeared in seven environments, and in four out of seven environments, the TGW QTL were localized on the TaGSr-4B locus and showed significant contributions to the phenotype. Based on the marker, two allele groups of Westonia and Kauz formed showed significant differences on TGW in eight environments. In agreement with the roles of GS genes on nitrogen and carbon remobilizations, TaGSr-4B is likely the candidate gene of the TGW QTL on 4B and the TaGSr-4B gene marker is ready for wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum) is one of the most important cereal crops and supplies food for 40% of human population (Pang et al. 2020). Due to the world's growing population, a high increase in wheat yield is required (Shiferaw et al. 2013). In general, wheat grain yield components include thousand grain weight (TGW), grain number per spike (GN) and spike number per area (Brinton and Uauy 2019). Among those, TGW showed high heritability, indicating the possibility of trait enhancement via genetic manipulation (Zhang et al. 2021). Several TGW QTL have been identified and potential candidate genes were revealed (Mohler et al. 2016; Xu et al. 2019; Guan et al. 2020; Wen et al. 2022).
In our recent study, 11 TGW QTL were detected on 1A, 1B, 4A, 4B, 4D, 5A, 5B, 7A, and 7B. Except 4D, 5A, 7A and 7B, all other QTL were detected in multiple field conditions or replicates (Zhang et al. 2021). In the literature, the TGW QTL was also reported on 1A (Arif et al. 2021), 1BL (Ren et al. 2021), 2DL (Li et al. 2021), 3A (Arif et al. 2021), 3D (Cui et al. 2014), 4A (Guan et al. 2018), 4BS (Guan et al. 2018; Xu et al. 2019; Li et al. 2021), 4BL (Guan et al. 2018; Xiong et al. 2021), 4D (Li et al. 2018), 5AL (Li et al. 2021), 5B (Yang et al. 2020), 5D (Li et al. 2018), 6A and 6B (Li et al. 2018; Arif et al. 2021), 6D (Cui et al. 2014), 7A and 7B (Guan et al. 2018; Li et al. 2018; Arif et al. 2021; Corsi et al. 2021), and 7D (Chen et al. 2020).
In addition, a number of genes for the seed size and TGW were identified, such as 1) a nuclear-encoded chloroplast protein gene Tabas1-B1 (Tabas1-B1a and Tabas1-B1b) on 2BL (Zhu et al. 2016); 2) grain size related gene—TaGS5-3A (Ma et al. 2016), TaGS-D1 (7D: 6.4 Mb bp) (Zhang et al. 2014b), TaTGW6-4AL (Hu et al. 2016a), and TaTGW-7A (Hu et al. 2016b); 3) a stress association protein (SAP) gene—TaSAP1-A1 (7A: 585.0 Mb) and TaSAP1-7B (7B: 544.4 Mb) (Chang et al. 2013).
Previous studies have demonstrated that the grain size and grain yield were directly influenced by the levels of stem water soluble carbohydrate (WSC) and the WSC remobilization efficiency (Dreccer et al. 2009; Mir et al. 2012). The stem WSC levels before anthesis were associated with 57 and 74% of the grain yield of wheat and barley, respectively (Gallagher et al. 1976). On the stem WSC remobilization pathway, a fructan 1-exohydrolase (1-FEH w3) allele on 6B contributed to high TGW under drought conditions (Zhang et al. 2015).
A carbon consumer glutamine synthetase (GS) is considered as a connection point between the organic and inorganic phases, and a regulator between nitrogen and the carbon cycles in the chloroplast (Németh et al. 2018). The increasing amounts of carbon skeleton additional subunits lead to an increasing efficiency of GS2 oligomer (Németh et al. 2018). The expression levels of GS were regulated by the levels of nitrogen (N) and carbon (C) metabolites, in particular, controlled by the relative abundance of carbon skeletons (Oliveira and Coruzzi 1999). In their study, the transcription of GS1 and GS2 in cytosolic and chloroplast, respectively, was mediated and induced by light and sucrose level. The level of sucrose also changes the level of GS enzyme activity. GS in wheat was classified into four families: TaGS1, TaGS2, TaGSr and TaGSe (Bernard et al. 2008; Yin et al. 2022). Based on their subcellular locations, TaGS1, TaGSr and TaGSe belong to the cytosolic isoform while TaGS2 in the plastidic isoform (Swarbreck et al. 2011). During the leaf senescence in grain filling stage, the cytosolic isoform (GS1 and GSr) were predominant which suggested that they were critical for nitrogen remobilization to grain (Bernard et al. 2008). According to phylogenetic tree analysis of four GS clusters of GS1;1, GS1;2, GS1;3 and GS2, TaGS1 was classified to cluster GS1;1, TaGSr to GS1;2, TaGSe to GS1;3, and TaGS2 to GS2 (Thomsen et al. 2014). In addition to its function in ammonia assimilation, GS is critical for high grain production (Németh et al. 2018). In barley, the grain yield and nitrogen use efficiency (NUE) were improved through the over expression of HvGS1;1 (Gao et al. 2019). In wheat, the grain number per spike and TGW were increased with the high TaGS2-2Ab gene expression level in the transgenic lines (Hu et al. 2018). Yu et al. (2021) found that sulphur regulates the GS activity in wheat leaves to increase NUE and grain yield (Yu et al. 2021). TaGS2-A1, B1 and D1 were mapped on group 2 and GS-2B were suggested as a candidate gene for grain protein content (GPC) QTL on 2B (Li et al. 2011; Nigro et al. 2020). TaGS1 was mapped in group 6 chromosomes, while TaGSr and TaGSe were in group 4 (Bernard et al. 2008; Nigro et al. 2019; Wang et al. 2020).
The TGW QTL on 4B have been particularly highlighted since those are close to or overlapped with one of the green revolution genes—Rht1 gene (Duan et al. 2020; Guan et al. 2020; Wen et al. 2022). However, the TGW QTL in the population of Westonia × Kauz was localized beyond the Rht1 gene (Zhang et al. 2013). The QTL-hot spot for TGW was recently dissected into two QTL clusters (4B.1 and 4B.2) (Guan et al. 2020). The 4B.1 QTL was considered as the pleiotropic effects of Rht1, while the 4B.2 QTL seemed to form later in the grain filling stage although the candidate genes were not clear. Identifying the candidate genes of TGW QTL on 4B tends to be sophisticated but rather important for wheat yield improvement through breeding. The aim of the current study is to identify the candidate genes for TGW QTL on 4B.
Materials and methods
Plant materials
A wheat doubled haploid (DH) population of Westonia × Kauz with 225 DH lines were used for the TGW candidate gene identification. Westonia is a high yielding wheat cultivar bred in Western Australia. Kauz is a high yielding variety from the International Maize and Wheat Improvement Center (CIMMYT, EI Batan, Mexico) (Butler et al. 2005) and is drought tolerant (Rajaram et al. 2002). Both varieties achieve high yield with distinctive yield components, for example, the TGW in Westonia was significantly higher than Kauz, while the seed number per spike of Kauz is consistently higher than Westonia (Zhang et al. 2013). Kauz carries dwarfing gene—Rht1 (Rht-B1b and Rht-D1a) (Butler et al. 2005), while Westonia carries Rht2 gene (Rht-B1a and Rht-D1b) (Ellis et al. 2002; Botwright et al. 2005; Zhang et al. 2013). Those Rht genes were segregated in the Westonia × Kauz population and a TGW QTL was detected beyond the Rht1 gene locus (Zhang et al. 2013).
Glasshouse and field experiments
The previous TGW data of the 225 DH lines were included, which were from a glasshouse experiment at Murdoch University (Perth) in 2010, field experiments in Katanning in 2010 and Shenton Park in 2011. In glasshouse, three plants per line were grown in 4 L free-draining pots filled with potting mix prepared according to Zhang et al. (Zhang et al. 2009). The potting mix soil with sufficient fertilizer in each pot was weighed and pasteurized before planting. The pots were well-watered using mats underneath the pots and the pots were rotated on alternate days for receiving equal radiation exposure.
In Katanning (33.7° S, 117.6° E), due to the seed availability, the parental and 220 DH lines were grown under rain-fed condition and each line was in a separate 4.5 m2 (1.5 × 3 m) plot. In 2010, the annual rain fall in Katanning was low as 246 mm while the average was 437 mm from 1999 to 2012. In particular, during the grain filling period (August–October, 2010), the total rain fall was 27 mm while the average was 135 mm. Crops were managed with adequate nutrition, and spraying of herbicides and pesticides to control weeds and leaf diseases, respectively. In 2011, the parental and 223 DH lines were grown under adequate nutrition and water level (Zhang 2008) at Shenton Park field station (Perth, 32.0° S 115.9° E). The plot size for each line was 1.2 m2 (1.5 × 0.8 m). No spray was applied, and weeds were manually removed.
Plant height of three individual or representative (field) plants was measured at maturity. In glasshouse, all spikes (3) of each plant were taken for GN and TGW measurements while 20 and 3 random spikes from each DH line were taken in Shenton Park and Katanning field, respectively. The GN per spike and TGW were determined as grain parameters.
In 2016, two field experiments were carried out in Shenton Park field station and Wongan Hills (30.9° S, 116.7° E) (Zhao 2019). Complete randomised block designs were used for the two field sites. In Shenton Park, two parental lines and 152 DH lines were chosen for trials based on the seed availability. In addition to the same level base fertilizer, three nitrogen treatments (0, 50, 100 kg/Ha) were applied. Half of the 152 DH lines (71 lines) were replicated for final spatial adjustment. The individual plot size was 0.67 m2 (1 × 0.67 m). The nitrogen fertilizer was applied at the stem elongation stage. In Wongan Hills, the same nitrogen treatments (0, 50, 100 kg/Ha) were applied to the field trial. A group of 143 DH lines and the two parental lines were grown with individual plot size of 1.2 m2 (2 × 0.6 m). The nitrogen rate was used as a blocking factor in the design for those two field trials. The pesticides were sprayed in both stations and weeds were removed manually. During harvesting, five uniform plants from each plot were selected for phenotyping the yield components. For QTL analysis, each nitrogen rate was treated as one field condition, meaning a total of six field conditions in both field sites were evaluated.
Linkage map construction
The 90 K SNPs were used to enrich the existing genetic linkage map that was constructed using SSR, 9 K SNPs and allele specific markers of Rht-B1b, Rht-D1b, Vrn-A1a, Vrn-B1a, and Vrn-D1a (Zhang et al. 2014a), etc. The SNP markers with large missing values of genotyping (> 25%) were excluded together with the distorted markers and double-cross markers. Most co-segregating markers were made redundant and removed from the genetic map. A fine map of Westonia and Kauz (WK) population was constructed with 3798 markers using Map Manager (Manly et al. 2001) and the QTL mapping package R/qtl (Broman and Sen 2009).
QTL mapping
Inclusive composite interval mapping (ICIM) (IciMapping V4.1; http://www.isbreeding.net) was used for QTL detection (Li et al. 2015). The inclusive composite interval mapping addition (ICIM-ADD) method was selected for QTL mapping (Li et al. 2007). The mean value of replicates of individual DH line in each environment was used for QTL detection under a LOD score of 2.5. Permutations were set to 1000 at a significant level of 0.05.
RNAseq analysis
A field drought experiment was carried out in 2011 (Zhang et al. 2015). The main stems (sheath included) were used for RNA extraction and later RNAseq analysis (Zhang et al. 2015; Al-Sheikh Ahmed et al. 2018). Samples collected between two to five weeks after anthesis at weekly interval were used for the RNAseq data analysis for this study. Three biological replicates of RNA extracts in each treatment (drought and irrigated) and growth stages were used for cDNA library construction with the Illumina TruSeq RNA v2 protocol and then sequenced using the Illumina HiSeq platform (100 bp paired end reads) using standard protocols at the Australian Genome Research Facility. The sequence data were obtained through the Illumina CASAVA1.8 pipeline. The data quality was checked by FastQC, and the low-quality reads were trimmed. Using Biokanga (version 3.4.2), those sequences were then aligned to the complete CDS contigs from the wheat sequencing consortium (https://www.wheatgenome.org/). Different gene expression analysis was conducted using DEseq (version 1.12.1) (Anders and Huber 2010) and edgeR (version 3.2.4) (Robinson et al. 2009). Gene expression level was normalized using model-based scaling normalization or Trimmed Mean of M-values (TMM).
Database searches and sequence analysis
The gene sequences are available from the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and Ensembl Plants (https://plants.ensembl.org). These websites were used for a query to obtain closely related wheat genome sequences. The International Wheat Genome Sequencing Consortium (IWGSC) Survey Sequence Repository was used for exploring the upstream and downstream regions of the respective genes. The retrieved DNA sequences were analysed using CLC genomics workbench 10 and Geneious V.7.1.9.
Primer design, PCR amplification of genomic DNA, cloning and enzyme digestion
Primer sequences were designed based on the reference sequence from IWGSC. The PCR amplification and analyses followed the previous protocol (Zhang et al. 2008). PCR products were centrifuged down at a high speed (14,000 RPM) in a filter tube and sequenced directly (ABI 377, Applied Biosystems). The enzyme digestion was performed in a total reaction volume of 20 μl that included 10 μl of PCR products, 10 units of enzyme (1 μl), and 1× rCutSmartTM buffer (2 μl). The mixture was incubated at 37 °C overnight. The digested PCR products were run in a 1.5% agarose gel.
Statistical analysis
Phenotypic and gene expression data were analysed by multivariate analysis of variance (MANOVA) using the general linear model implemented in IBM SPSS statistics 24 (https://www.ibm.com/au-en/products/spss-statistics). Post hoc Tukey’s Multiple Range tests were used to identify significant groupings. T test was used for the significance of parental phenotypes and different genotypic allele groups. The box and violin plots were generated through the ggplot2 package in R. The broad-sense heritability was calculated through Package lme4 in R-studio.
Results
Phenotypic variations
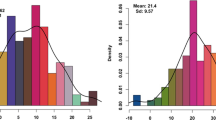
Wheat cultivars of Westonia and Kauz in past studies consistently showed significant differences in grain yield components, such as Westonia with significantly higher TGW, while Kauz with significantly higher GN (Zhang et al. 2013) (Supplementary Table S1). The plant height of Westonia was significantly higher than Kauz in glasshouse. Since the TGW QTL with high F-value (19.7–37.9) were detected on 4B and strongly linked with the Rht1 gene locus, further TGW QTL analysis was performed within this study for candidate gene identification. Apart from the previous data obtained in glasshouse and Katanning field station in 2010, and Shenton Park in 2011, a subset of 152 and 143 DH lines were planted in two field trials of Shenton Park and Wongan Hills in 2016, respectively. On average, the TGW was lower in comparison with previous data in glasshouse and Shenton Park, slightly higher than that from Katanning (Fig. 1). In 2016, slight differences were appeared between different nitrogen levels. In Shenton Park, TGW increased along with the N levels, while in Wongan Hills, it presented a different trend, which may be due to the different soil conditions. Overall, all TGW data showed normal distributions (Fig. 1 and Table S2). The TGW in nine environments showed the highest heritability (0.88) in comparison with the other phenotypes. In parallel, the TGW associated phenotypes of plant height, grain weight (GW), grain number per spike in glasshouse, Katanning (2010) and Shenton Park (2011) also showed normal distributions (Fig. S1).
The distribution of thousand grain weight (TGW) in the DH population of Westonia and Kauz in nine environments. GH: glasshouse in 2010; Ka: Katanning in 2010; SH: Shenton Park in 2011; SH0, SH50, SH100: Nitrogen (N) level 0, 50 and 100 kg/ha in Shenton Park in 2016; WH0, WH50, WH100: N level 0, 50 and 100 kg/ha in Wongan Hills in 2016
Genetic map of WK population
A fine map was constructed for QTL analysis. A total of 3798 markers formed 2651 bin markers and were allocated into 21 linkage groups with the total genome coverage of 7423.4 cM and an average bin marker interval of 2.8 cM. The total map length for each chromosome ranged from 202.1 cM (6D) to 617.6 cM (7A) and the minimum average bin marker interval was on 2B (1.7 cM) and maximum on 7D (12.4 cM) (Table S3).
TGW QTL on 4B
A TGW QTL cluster was detected with three QTL linked together on positions 125–133, 135–137 and 141–143 cM. The QTL cluster was 40 cM beyond Rht1. The positive effects of all QTL were contributed by Westonia with the highest LOD value of 13.4 and the largest PVE value of 21.2% on the position of 132–137 cM (Fig. 2 and Table S4).
The QTL of thousand grain weight (TGW) detected on 4B in six environments together with the QTL of grain weight (GW) per spike, grain number (GN) per spike, plant height (Height) in glasshouse, Katanning, and Shenton Park. A TGW QTL cluster highlighted in light blue. GH: glasshouse in 2010; Ka: Katanning in 2010; SH: Shenton Park in 2011; sh0, sh50, sh100: Nitrogen (N) level 0, 50 and 100 kg/ha in Shenton Park in 2016; wh0, wh100: N level 0 and 100 kg/ha in Wongan Hills in 2016
The QTL for GW and plant height were on the Rht1 gene region while a grain number (GN) QTL was at 24 cM beyond the Rht1 locus and another plant height QTL was overlapped with the TGW QTL cluster (Fig. 2 and Table S4).
Candidate genes within TGW QTL cluster on 4B
Within the TGW QTL cluster on 4B, there were 773 genes (TraesCS4B02G236800–TraesCS4B02G371300) between markers gwm192b and barc163 (4B: 460–580 Mb in the physical map) (Table S5). Since the grain weight was accumulated through the grain filling stage, and the stem WSC was heavily involved in grain filling, the stem RNAseq data between 11 and 31 days after anthesis (DAA) were used for candidate gene selection. Out of the 773 genes, 315 gene expression data were captured, and four genes showed two transcripts in Kauz but one in Westonia. Those genes were TraesCS4B02G240100 (4B: 497.86 Mb), TraesCS4B02G240900 (4B: 499.90 Mb), TraesCS4B02G251800 (517.95 Mb), and TraesCS4B02G262900 (532.72 Mb). The first gene—TraesCS4B02G240100 was described as two-component response regulator in Ensembl Plants (ENBK-EBI) since it contained Myb (a member of myeloblastosis family) domain and a receiver domain—signal transduction response regulator. The second gene—TraesCS4B02G240900 was a glutamine synthetase (GS) on 4B. GS is an important gene for nitrogen (NH4+) absorption and involved in glutamine synthase for protein formation in plants. The third gene—TraesCS4B02G251800 is a putative Nodulin-related protein and belongs to transmembrane protein family (Denancé et al. 2014). Overexpression of the Nodulin-related protein 1 decreased the thermotolerance through reducing the accumulation of abscisic acid (ABA) after heat shock treatment in Arabidopsis (Fu et al. 2010). The fourth gene—TraesCS4B02G262900 is a putative mannitol transporter. The high gene expression levels and mannitol transport activity of mannitol transporter are parallel with the salt and drought-tolerant capacity in olive trees (Conde et al. 2011). The salt and drought levels significantly increased the mannitol transport activity and mannitol transporter gene expression level in wheat (Abebe et al. 2003).
From the comparisons of the gene expression levels, no significant differentiation in gene expression levels between Westonia and Kauz was detected on TraesCS4B02G251800 (517.95 Mb) while the only difference showed between treatments at 25 DAA in Westonia on TraesCS4B02G240100 (497.87 Mb) (Fig. S2 and Table S6). The gene expression of TraesCS4B02G262900 gradually decreased after 17 DAA at irrigated condition in both varieties. The significantly lower gene expression levels were observed under drought relative to irrigated in both varieties after 17 DAA while high small sugar molecules (glucose, fructose and sucrose) significantly increased under drought (Zhang et al. 2015). In addition, no significant difference was presented between Westonia and Kauz in both treatments. Those results indicate that gene—TraesCS4B02G262900 may not contribute significantly to the sugar remobilization. However, the gene expression of TaGS-4B (TraesCS4B02G240900, 499.90 Mb) gradually increased and was significantly enhanced under drought, especially in Westonia (Fig. 3). It tends to be a candidate gene for the TGW QTL.
Different gene expression levels of two TraesCS4B02G240900 transcripts between Westonia and Kauz in different growth stages (days after anthesis) and treatments. K: Kauz; Wes: Westonia; D: drought; W or WW: well-watered. The vertical bars represent SE; values with the same letter are not different at p < 0.05
TaGS-4B gene expression levels
There were two GS transcripts in Kauz, while only one transcript showed in Westonia (Fig. 3 and Table S6). Low expression levels ranged 0–8 units were showed on TraesCS4B02G240900.1 and no significant difference was identified between treatments. However, the expression levels on TraesCS4B02G240900.2 ranged 95–1563 units and displayed clear increasing patterns in both treatments and varieties along with the growth stages, especially under drought. At 31 DAA, the expression level was significantly higher under drought compared to the well-watered in Westonia, while the same pattern also appeared in Kauz but with non-significance (Fig. 3). The results indicate that GS may play a role in nitrogen remobilization efficiency under drought.
TaGS-4B is TaGSr1
It was reported that there were four groups of GS in wheat, including TaGS1, TaGS2, TaGSr and TaGSe with three copies of TaGS1 and TaGS2, two copies of both TaGSr and TaGSe while both TaGSr and TaGSe were on group 4 chromosomes (Bernard et al. 2008). The accession IDs of AY491968 (TaGSr1), AY491969 (TaGSr2), AY491970 (TaGSe1) and AY491971 (TaGSe2) in NCBI were used to blast against the wheat genome. TaGS-4B (TraesCS4B02G240900) showed 100% identity with TaGSr1, while TaGS-4D (TraesCS4D02G240700) was identical to TaGSr2 (Fig. 4a). Another homologous gene on 4A was identified as TraesCS4A02G063800. All those TaGSr genes have the exact same size of nine exons and eight introns. For clear identification, those TaGSr genes have been named as TaGSr-4A, TaGSr-4B and TaGSr-4D (Fig. 4b). Similarly, three homologous genes of TaGSe were also identified in group 4 chromosomes, including TraesCS4B02G047400 and TraesCS4D02G047400 standing for TaGSe1 and TaGSe2, and the later—TraesCS4A02G266900 unidentified before (Fig. 4a). For clear identification, those TaGSe genes have been named as TaGSe-4A, 4B and 4D. TaGSe-4A and 4D showed 12 exons and 11 introns, while TaGSe-4B had one less exon and intron as the exon 7 and 8 were combined as exon 7 (Fig. 4b). TaGSe-4B and TaGSr-4B were located on the short and long arms on 4B, respectively.
Polymorphisms on TaGSr-4B
From the gene expression analysis within the TGW QTL cluster, TaGSr-4B tends to be a candidate gene for the QTL. The GS gene sequences including the promoter regions and downstream untranslated regions (UTR) were isolated from Westonia and Kauz using primer pairs designed specific for 4B chromosome (Table S7 and Fig. S3). In the promoter region, one bp insertion on the position of 268 bp, two SNPs on the positions of 195 and 221 bp in Westonia ahead of the coding region were identified. One and two SNPs were detected on intron 1 and 3′ terminal UTR region, respectively. Moreover, a 13-bp deletion was revealed on the 3′ terminal UTR region in Westonia (Fig. 5a). Interestingly, all above SNPs, a 1-bp insertion and a 13-bp deletion occurred in Westonia only while the sequences of Kauz were the same as Chinese Spring.
Gene sequence polymorphisms on TaGSr-4B between Westonia and Kauz. a The one SNP insertion (− 268 bp) and two SNPs (− 221 bp and − 195 bp) in the promoter region, and two SNPs (+ 372 bp, + 654 bp) and a 13-bp deletions on the 3′ untranslated regions, and one SNP in intron 1 (the related four motifs were listed); b. the cleaved amplified polymorphic (CAP) marker showed 102 bp difference between Westonia (308 bp) and Kauz (206 bp) using cutting enzyme –BcoDI (GTCTCN’/CAGAG) on − 221 bp in the promoter. GA gibberellin, MYB myeloblastosis, TALE transcription activator-like effector
Within the polymorphisms between parental lines, on the promoter region (− 221 bp), the sequence differences involved two different motifs including water stress motif—TAACTG in Kauz and gibberellin (GA) motif—TAACAGA in Westonia. On the 3′ terminal UTR region, the SNP mutation (+ 372 bp) in Westonia led to an additional motif of MYB while the 13-bp deletion in Westonia caused a motif—transcription activator-like effector (TALE) deleted.
Based on those SNPs between Westonia and Kauz, a primer pair of En760F and En1045R was designed since the SNP on the position of − 221 bp aligned with the enzyme cutting sequence of BcoDI/BsmAI (GTCTCN’/CAGAG; detected with NEBcutter V2.0 software). The primer pair was used for amplifying a 308 bp fragment in both varieties. The fragment in Kauz was cut into two fragments of 101 and 206 bp, while it remained the same size in Westonia (308 bp). The 102 bp fragment difference between Westonia (308 bp) and Kauz (206 bp) was easily identified in a 1.5% agarose gel (Fig. 5b). This cleaved amplified polymorphic (CAP) marker was used for screening TaGSr-4B in the WK DH population, which was mapped above an SSR marker WMC657 (Fig. 6).
The QTL of thousand grain weight (TGW) detected on 4B in seven environments together with the QTL of grain weight (GW) per spike, grain number (GN) per spike, plant height (Height) in glasshouse, Katanning, and Shenton Park while TaGSr-4B gene marker (GS_4B) was included in the map. TaGSr-4B gene (GS_4B) was mapped above the SSR marker—WMC657, and the TGW QTL cluster highlighted in light blue were on/near the region of the gene marker GS_4B (TaGSr-4B). GH: glasshouse in 2010; Ka: Katanning in 2010; SH: Shenton Park in 2011; sh0, sh50, sh100: Nitrogen (N) level 0, 50 and 100 kg/ha in Shenton Park in 2016; wh0, wh100: N level 0 and 100 kg/ha in Wongan Hills in 2016
New TGW QTL on 4B
The WK genetic map including TaGSr-4B CAP marker (GS_4B) was used for TGW QTL reanalysis on 4B. Interestingly, a TGW QTL was detected in one more environment—the Shenton Park field trial in 2010 (Shenton2010). Under four field conditions (TGW_SH, TGW_sh0, TGW_sh50, TGW_sh100), the TGW QTL were laid on the GS_4B marker region while no change was noticed on TGW QTL in glasshouse. The TGW QTL in N0 and N100 in Wangan Hills were moved to 10 and 7 cM below in comparison with the previous QTL, respectively (Fig. 6 and Table S8). The positive allele of all TGW QTL were from Westonia accounting for 8–25% of the phenotype variations.
Since the Rht1 gene was reported to contribute to TGW (Guan et al. 2020), its role for TGW in the WK population was further analysed using plant height. The same results appeared that the QTL of plant height in three environments were on the Rht1 gene region which was 40 cM above the TGW QTL cluster. It could conclude that the Rht1 gene that confers the plant height may not contribute to the TGW QTL in the current population (Fig. 6). One height QTL (LOD: 12.4) attributed to Westonia was overlapped with the TGW QTL (Fig. 6 and Table S8), indicating the overall contribution of plant height to TGW (Wen et al. 2022).
The CAP marker of TaGSr-4B correlates with superior TGW
The TaGSr-4B marker—GS_4B was used to separate the WK population into two allelic groups of Westonia and Kauz. Significant differences between the two groups were identified in TGW under eight environments. The Westonia type increased TGW by 5–12% in comparison with the Kauz type (Fig. 7).
Significant differences on thousand grain weight between Westonia and Kauz allele types in DH population under eight environments. GH: glasshouse in 2010; Ka: Katanning in 2010; SH: Shenton Park in 2011; SH0, SH50, SH100: Nitrogen (N) level 0, 50 and 100 kg/ha in Shenton Park in 2016; WH0, WH50, WH100: Nitrogen (N) level 0, 50 and 100 kg/ha in Wongan Hills in 2016
Discussion
Effects of multiple transcripts
Genetic variants influencing RNA processing are similar to the variants affecting transcription. The sequence variations on pre-mRNA would impact the stability, translation and structure of mRNAs, causing phenotypic diversity (Gingeras 2007; Manning and Cooper 2017). Apart from the basal RNA processing, some genes have alternative splicing and express multiple transcripts, which may encode different isoforms. The major differential pro-mRNA processing is mediated in a specific tissue and stage, or in response to particular regulators (Manning and Cooper 2017). In this study, the genes within the TGW QTL cluster with multiple transcripts were chosen for their potential function for the phenotypes.
A total of four genes including TaGSr-4B within the TGW QTL cluster were detected with two transcripts in Kauz and one in Westonia. TaGSr-4B showed gradually increased gene expression patterns along with the growth stages. The gene expression levels of TaGSr-4B were almost in parallel with the sucrose and fructose level measured previously (Zhang et al. 2015). The significant differences in gene expression levels were presented between treatments in Westonia. In addition, GS gene was previously reported to be associated with TGW in Maize (Hirel et al. 2001; Martin et al. 2006) and wheat (Habash et al. 2001; Hu et al. 2018). Therefore, TaGSr-4B was chosen for this study.
The associations between GS genes and TGW
Several grain yield-related QTL showed linkages with the localization of GS. For example, in maize, the QTL of grain yield and TGW were colocalized on the region of GS on chromosome 5 (Hirel et al. 2001). Strong positive correlations between grain yield and GS activity were also observed (Gallais and Hirel 2004). Further knockout ZmGln1;3 and ZmGln1;4, the kernel number and kernel weight were reduced in the mutants (Martin et al. 2006). In rice, the QTL of one-spikelet weight was on the region of OsGS1, and the OsGS1;1 knockout mutant showed a severe delay in grain filling under normal nitrogen level (Obara et al. 2001; Tabuchi et al. 2005). In barley, overexpression of HvGS1;1 achieved higher grain yield and NUE compared with the wild-type under three nitrogen levels (Gao et al. 2019). In sorghum, the transgenic lines with higher levels of a sorghum cytosolic GS gene—Gln1 transcripts exhibited a higher GS activity and showed 2.1-fold increased shoot biomass in relative to controls (Urriola and Rathore 2015). In wheat, the transgenic lines with favourable allele—TaGS2-2Ab increased GS activity and enhanced grain yield, grain number per spike and TGW (Hu et al. 2018). During senescence, the increased GS1 activity in the transgenic wheat lines enhanced the capacity of nitrogen accumulation in grain resulting in increased root and grain dry weight (Habash et al. 2001). Those results conclude that high gene expression levels of GS lead to increased GS activities which results in high grain yield and TGW. From our RNAseq results, the transcription levels of TaGSr-4B were gradually increased along with the growth stages. In particular, the gene expression levels were significantly higher under drought in Westonia, which evidence the higher remobilization during the late grain filling stage leading to the significantly high TGW.
In addition, the TGW QTL on 4B has been highlighted by a number of researchers (Li et al. 2018, 2021; Xu et al. 2019; Arif et al. 2021; Corsi et al. 2021). In their studies, the TGW QTL was overlapped or very close to the Rht1 locus, or plant height QTL (Li et al. 2018; Xu et al. 2019; Guan et al. 2020; Wen et al. 2022). However, in our previous study using the population of Westonia and Kauz, the TGW QTL was located beyond the polymorphic Rht1 locus (Zhang et al. 2013). So far, the map of Westonia and Kauz was enriched using 90 K SNPs. The bin markers on 4B have been increased from 28 to 87. The TGW QTL were clearly showing 40 cM beyond the Rht1 locus. Therefore, it is unlikely to state that Rht1 had the major contribution to the TGW QTL in the WK population although the association still exists due to the linkage on 4B. In fact, from the QTL and TaGSr-4B allele analysis, TaGSr-4B is likely the major candidate gene for the TGW QTL in the population of Westonia and Kauz. Nevertheless, a possibility still exists in that both TaGSr-4B and Rht1 contribute to TGW but the TaGSr-4B has a much stronger effect that masks the Rht1 effect in the WK population.
GS family and function
Glutamine synthetase (GS; EC 6.3.1.2) is one of the main enzymes accumulating inorganic N through the ATP-dependent conversion from glutamine into glutamate (Yin et al. 2022). GS has been focused by multiple studies on nitrogen use efficiency in plants (Hirel et al. 2001; Zhang et al. 2017; Hu et al. 2018). High GS activities were correlated well with high grain yield due to its important role in N-remobilization (Gallais and Hirel 2004). High N-remobilization after flowering can also affect the grain filling leading to high grain weight (Gallais and Hirel 2004). A particular study on the interdependence of C and N metabolism in maize kernel development revealed that the supply of N assimilates facilitates kernel’s utilization of carbohydrates (Below et al. 2000). The expression levels of GS were controlled by the relative abundance of carbon skeletons, and the level of GS enzyme activity is regulated by the level of sucrose (Oliveira and Coruzzi 1999).
The sequences of GS isoforms include a range of light-responsive elements, auxin-responsive elements and binding sits for transcription factors such as MYB and Dof (Castro-Rodríguez et al. 2011). MYB transcription factor Circadian Clock-Associated 1 (CCA1) in Arabidopsis was predicted to have a role in regulating AtGln1;3 expression under the availability of C skeletons (Gutiérrez et al. 2008; Gutiérrez 2012). It was suggested that transcription factor—DNA-binding One Zinc Finger (Dof)1 was involved in the regulation of C4 photosynthetic phosphoenolpyruvate carboxylase (C4PEPC) gene expression, and Dof2 mediated multiple gene expressions associated with carbon metabolism pathway (Yanagisawa 2000, 2002).
From KEGG analysis, TaGSr-4B is involved in the glyoxylate and dicarboxylate metabolisms (path ko00630), energy metabolism (nitrogen metabolism, path: ko00910), amino acid metabolism (alanine, aspartate and glutamate metabolisms, path: ko00250), and arginine biosynthesis (path: ko00220). In addition, TaGSr-4B is involved in environmental signal transduction (two-component system, path: ko02020) and signaling and cellular processes (exosome, brite: ko04147), as well as in cell growth and death (necroptosis, path: ko04217). More importantly, the substrate-glutamate in glutamine synthesis is formed from alpha-ketoglutarate generated from sucrose in mitochondria. The metabolic pathways clearly indicates associations between the carbon levels and N assimilation. Taken together, GS gene expression is regulated by the available C skeleton and amino acid and involved in the interdependence of N and C metabolisms.
There have been two types of GS (GSI and GSII) identified in plants, with the genome sequence size of GSI larger than GSII (Swarbreck et al. 2011). The GSI is in the cytosol, while the GSII encoding the plastidic isoform is located and active in mainly chloroplast and merely mitochondria (Taira et al. 2004). In wheat, the cytosolic GSI have three subtypes (TaGS1, TaGSr and TaGSe). During the grain development, TaGS1 and TaGSr were the predominant forms presented in root, flag leaf, peduncle and glume, in particular, TaGSr showed high gene expression in peduncle (Bernard et al. 2008). During leaf senescence and grain filling, the mesophyll cells disappeared, while the vascular cells containing TaGS1 (in the vascular tissue) and TaGSr (in the phloem sieve elements) remained intact, which further support the roles of assimilate remobilization (Bernard et al. 2008).
For the mRNA stability and translation efficiency, untranslated regions (UTRs) of genes are one of the crucial regulator (Rao et al. 2017). In comparison with the TaGSr-4B sequences of Westonia and Kauz, no difference was found in the coding region. However, two SNPs and a 1-bp insertion in Westonia were identified in 5′-UTR while two SNPs and a 13-bp deletion in Westonia in the 3′-UTR region. In addition, one SNP was identified in Intron 1. Particularly, at the position of − 195 bp, one motif (TAACTG) in Kauz was related to water stress, whereas the one (TAACAGA) in Westonia was a gibberellin (GA) responsive motif. The Kauz motif (TAACTG) found in the simian virus 40 enhancer and the maize bronze-1 promoter is a consensus MYB recognition sequence bound by ATMYB2 (Urao et al. 1993), which was predicted as a transcriptional activator in ABA-inducible expression of a dehydration-responsive gene, rd22, therefore, involved in regulation of genes that are responsive to water stress in Arabidopsis (Abe et al. 1997). It was suggested that genes with a MYB transcription factor (TF) binding motif in their promoters were controlled by MYB TF, which was induced by ABA (Lewis et al. 2015). Coincidently, a GA responsive element was on the same promoter region. GA is a plant hormone in regulating of plant growth. The Westonia motif TAACAGA is a GA response complex which is conserved in the promoters of alpha-amylase and proteinases in cereals (Sutoh and Yamauchi 2003). In addition, there was a MYB binding site (AACCAAAC) fitted MYB binding motif MACCWAMC ([AC]ACCWA[AC]C) in the 3′-UTR region followed by a 13-bp deletion in the Westonia type TaGSr-4B. Within the 13-bp deletion, one motif of TALE was missed. The 3′-UTR region plays a crucial role in gene expression through influencing the mRNA localization, stability, export, and translation efficiency and also establishes protein–protein interactions (Mayr 2019). TALE transcription factors could adjust the gene expression and also be a N-terminal translocation signal or a signal for unique chromosome targeted (Morbitzer et al. 2010).
The contributions of other genes
While the TGW QTL in the most environments were overlapped with the TaGSr-4B locus, the TGW QTL in glasshouse, and in N0 and N100 field trials in Wongan Hills are still 5 and 13 cM beyond the TaGSr-4B locus, respectively. Although the TaGSr-4B locus has been confirmed with the highest stringent in conferring the TGW phenotype, it is still possible that some other genes within the QTL also contribute to the phenotype. For further identifying other potential candidate genes, the entire gene expression profile on 4BL was drawn from the RNAseq data of Westonia and Kauz, and genes within the TGW QTL cluster (460–580 Mb) were investigated. Besides the multiple transcripts, the potential candidate gene selections followed the criteria: (1) the maximum expression level is over 100; (2) the average differentiation expression between Westonia and Kauz is over four times. As a result, two potential candidate genes were selected, including TraesCS4B02G222300 (466.96 Mb) and TraesCS4B02G225400 (471.96 Mb) (Fig. S4). The average expression levels of TraesCS4B02G222300 were 12.7 and 161.1 in Kauz and Westonia, respectively, with the differentiation of 12.7-fold (Fig. S4). The gene function is related to the transcription initiation factor IIF, alpha subunit (TFIIF). TFIIF is a transcription regulator and promotes transcription elongation (Gaudet et al. 2011). The different expression levels between Westonia and Kauz may regulate the gene expression in the network. TraesCS4B02G225400 is a heat shock protein with the average gene expression level of 130.3 in Westonia, whereas no gene expression was detected in Kauz (Fig. S4). Heat shock proteins are the key components responsible for protein folding, assembly, translocation, and degradation in normal cellular processes or under stress conditions (Park and Seo 2015). Further investigations are required for those potential candidate genes in contributing to the TGW QTL.
Conclusion
TGW is one of the three components of grain yield in wheat. The candidate gene identification of TGW QTL on 4B is crucial for its application in wheat breeding. A TGW QTL with a high F-value (37.9) was identified in a large interval on 4B in the DH population of Westonia × Kauz. Further wheat 90 K SNP array was screened across the DH population, and a highly saturated map was generated. The TGW data of nine environments formed a robust TGW QTL cluster including 773 genes. Within the QTL cluster, the gene expression data were derived from a RNAseq data of Westonia and Kauz. After filtering the differential gene expression levels and multiple transcripts, TaGSr-4B was selected as a potential candidate gene for the TGW QTL. The sequence variations were identified between Westonia and Kauz, and a CAP marker was generated for the gene mapping and the TGW QTL reanalysing. Surprisingly, in four out of seven environments, the TGW QTL were localized on the TaGSr-4B locus and showed significant contributions to the phenotype. The marker was used to separate the Westonia and Kauz allele types. Significant allelic effects on TGW were detected in eight out of nine environments. In agreement with the roles of GS genes on nitrogen remobilization in parallel with the functions of carbon remobilization, TaGSr-4B is most likely the candidate gene of the TGW QTL on 4B in Westonia and Kauz population. The TaGSr-4B gene marker can be used in wheat breeding. Since there were three TGW QTL were mapped in slightly different locations, it cannot rule out that other genes may also contribute to TGW.
Data availability
Electronic supplementary material The online version of this article (https://doi.org/xxxxx) contains supplementary material, which is available to authorized users.
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868. https://doi.org/10.1105/tpc.9.10.1859
Abebe T, Guenzi AC, Martin B, Cushman JC (2003) Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol 131:1748–1755. https://doi.org/10.1104/pp.102.003616
Al-Sheikh Ahmed S, Zhang J, Ma W, Dell B (2018) Contributions of TaSUTs to grain weight in wheat under drought. Plant Mol Biol 98:333–347. https://doi.org/10.1007/s11103-018-0782-1
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. https://doi.org/10.1186/gb-2010-11-10-r106
Arif MAR, Shokat S, Plieske J, Ganal M, Lohwasser U, Chesnokov YV, Kocherina NV, Kulwal P, Kumar N, McGuire PE, Sorrells ME, Qualset CO, Börner A (2021) A SNP-based genetic dissection of versatile traits in bread wheat (Triticum aestivum L.). Plant J 108:960–976. https://doi.org/10.1111/tpj.15407
Below FE, Cazetta JO, Seebauer JR (2000) Carbon/nitrogen interactions during ear and kernel development of maize. In: Physiology and Modeling Kernel Set in Maize. pp 15–24. https://doi.org/10.2135/cssaspecpub29.c2
Bernard SM, Møller ALB, Dionisio G, Kichey T, Jahn TP, Dubois F, Baudo M, Lopes MS, Tercé-Laforgue T, Foyer CH, Parry MAJ, Forde BG, Araus JL, Hirel B, Schjoerring JK, Habash DZ (2008) Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol Biol 67:89–105. https://doi.org/10.1007/s11103-008-9303-y
Botwright T, Rebetzke G, Condon A, Richards R (2005) Influence of the gibberellin-sensitive Rht8 dwarfing gene on leaf epidermal dell dimensions and early vigour in wheat (Triticum aestivum L.). Ann Bot 95:631–639
Brinton J, Uauy C (2019) A reductionist approach to dissecting grain weight and yield in wheat. J Integr Plant Biol 61:337–358. https://doi.org/10.1111/jipb.12741
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Statistics for biology and health. Springer, Dordrecht, Heidelberg
Butler JD, Byrne PF, Mohammadi V, Chapman PL, Haley SD (2005) Agronomic performance of Rht alleles in a spring wheat population across a range of moisture levels. Crop Sci 45:939–947
Castro-Rodríguez V, García-Gutiérrez A, Canales J, Avila C, Kirby EG, Cánovas FM (2011) The glutamine synthetase gene family in Populus. BMC Plant Biol 11:119. https://doi.org/10.1186/1471-2229-11-119
Chang J, Zhang J, Mao X, Li A, Jia J, Jing R (2013) Polymorphism of TaSAP1-A1 and its association with agronomic traits in wheat. Planta 237:1495–1508. https://doi.org/10.1007/s00425-013-1860-x
Chen Z, Cheng X, Chai L, Wang Z, Bian R, Li J, Zhao A, Xin M, Guo W, Hu Z, Peng H, Yao Y, Sun Q, Ni Z (2020) Dissection of genetic factors underlying grain size and fine mapping of QTgw.cau-7D in common wheat (Triticum aestivum L.). Theor Appl Genet 133:149–162. https://doi.org/10.1007/s00122-019-03447-5
Conde A, Silva P, Agasse A, Conde C, Gerós H (2011) Mannitol transport and mannitol dehydrogenase activities are coordinated in Olea europaea under salt and osmotic stresses. Plant Cell Physiol 52:1766–1775. https://doi.org/10.1093/pcp/pcr121
Corsi B, Obinu L, Zanella CM, Cutrupi S, Day R, Geyer M, Lillemo M, Lin M, Mazza L, Percival-Alwyn L, Stadlmeier M, Mohler V, Hartl L, Cockram J (2021) Identification of eight QTL controlling multiple yield components in a German multi-parental wheat population, including Rht24, WAPO-A1, WAPO-B1 and genetic loci on chromosomes 5A and 6A. Theor Appl Genet 134:1435–1454. https://doi.org/10.1007/s00122-021-03781-7
Cui F, Zhao C, Ding A, Li J, Wang L, Li X, Bao Y, Li J, Wang H (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659–675. https://doi.org/10.1007/s00122-013-2249-8
Denancé N, Szurek B, Noël LD (2014) Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol 55:469–474. https://doi.org/10.1093/pcp/pct198
Dreccer MF, van Herwaarden AF, Chapman SC (2009) Grain number and grain weight in wheat lines contrasting for stem water soluble carbohydrate concentration. Field Crop Res 112:43–54. https://doi.org/10.1016/j.fcr.2009.02.006
Duan X, Yu H, Ma W, Sun J, Zhao Y, Yang R, Ning T, Li Q, Liu Q, Guo T, Yan M, Tian J, Chen J (2020) A major and stable QTL controlling wheat thousand grain weight: identification, characterization, and CAPS marker development. Mol Breed 40:68. https://doi.org/10.1007/s11032-020-01147-3
Ellis ME, Spielmeyer WS, Gale KG, Rebetzke GR, Richards RR (2002) “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042. https://doi.org/10.1007/s00122-002-1048-4
Fu Q, Li S, Yu D (2010) Identification of an Arabidopsis Nodulin-related protein in heat stress. Mol Cells 29:77–84. https://doi.org/10.1007/s10059-010-0005-3
Gallagher JN, Biscoe PV, Hunter B (1976) Effects of drought on grain growth. Nature 264:541–542
Gallais A, Hirel B (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55:295–306. https://doi.org/10.1093/jxb/erh006
Gao Y, de Bang TC, Schjoerring JK (2019) Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol J 17:1209–1221. https://doi.org/10.1111/pbi.13046
Gaudet P, Livstone MS, Lewis SE, Thomas PD (2011) Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform 12:449–462. https://doi.org/10.1093/bib/bbr042
Gingeras TR (2007) Origin of phenotypes: Genes and transcripts. Genome Res 17:682–690. https://doi.org/10.1101/gr.6525007
Guan P, Lu L, Jia L, Kabir MR, Zhang J, Lan T, Zhao Y, Xin M, Hu Z, Yao Y, Ni Z, Sun Q, Peng H (2018) Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front Plant Sci. https://doi.org/10.3389/fpls.2018.00529
Guan P, Shen X, Mu Q, Wang Y, Wang X, Chen Y, Zhao Y, Chen X, Zhao A, Mao W, Guo Y, Xin M, Hu Z, Yao Y, Ni Z, Sun Q, Peng H (2020) Dissection and validation of a QTL cluster linked to Rht-B1 locus controlling grain weight in common wheat (Triticum aestivum L.) using near-isogenic lines. Theor Appl Genet 133:2639–2653. https://doi.org/10.1007/s00122-020-03622-z
Gutiérrez RA (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336:1673–1675. https://doi.org/10.1126/science.1217620
Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105:4939–4944. https://doi.org/10.1073/pnas.0800211105
Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA (2001) The role of cytosolic glutamine synthetase in wheat. Ann Appl Biol 138:83–89. https://doi.org/10.1111/j.1744-7348.2001.tb00087.x
Hirel B, Bertin P, Quilleré I, Bourdoncle W, Cl A, Dellay C, Al G, Cadiou S, Retailliau C, Falque M, Gallais A (2001) Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol 125:1258–1270. https://doi.org/10.1104/pp.125.3.1258
Hu MJ, Zhang HP, Cao JJ, Zhu XF, Wang SX, Jiang H, Wu ZY, Lu J, Chang C, Sun GL, Ma CX (2016) Characterization of an IAA-glucose hydrolase gene TaTGW6 associated with grain weight in common wheat (Triticum aestivum L.). Mol Breed 36:25. https://doi.org/10.1007/s11032-016-0449-z
Hu MJ, Zhang HP, Liu K, Cao JJ, Wang SX, Jiang H, Wu ZY, Lu J, Zhu XF, Xia XC, Sun GL, Ma CX, Chang C (2016b) Cloning and characterization of TaTGW-7A gene associated with grain weight in wheat via SLAF-seq-BSA. Front Plant Sci 7:1902–1902. https://doi.org/10.3389/fpls.2016.01902
Hu M, Zhao X, Liu Q, Hong X, Zhang W, Zhang Y, Sun L, Li H, Tong Y (2018) Transgenic expression of plastidic glutamine synthetase increases nitrogen uptake and yield in wheat. Plant Biotechnol J 16:1858–1867. https://doi.org/10.1111/pbi.12921
Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L, Kulasekaran S, Truman W, Littlejohn G, Prusinska J, Mead A, Steinbrenner J, Hickman R, Rand D, Wild DL, Ott S, Buchanan-Wollaston V, Smirnoff N, Beynon J, Denby K, Grant M (2015) Transcriptional dynamics driving mamp-triggered immunity and pathogen effector-mediated immunosuppression in arabidopsis leaves following infection with pseudomonas syringae pv tomato DC3000. Plant Cell 27:3038–3064. https://doi.org/10.1105/tpc.15.00471
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Li XP, Zhao XQ, He X, Zhao GY, Li B, Liu DC, Zhang AM, Zhang XY, Tong YP, Li ZS (2011) Haplotype analysis of the genes encoding glutamine synthetase plastic isoforms and their association with nitrogen-use- and yield-related traits in bread wheat. New Phytol 189:449–458. https://doi.org/10.1111/j.1469-8137.2010.03490.x
Li S, Wang J, Zhang L (2015) Inclusive composite interval mapping of QTL by environment interactions in biparental populations. PLoS ONE 10:e0132414–e0132414. https://doi.org/10.1371/journal.pone.0132414
Li F, Wen W, He Z, Liu J, Jin H, Cao S, Geng H, Yan J, Zhang P, Wan Y, Xia X (2018) Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor Appl Genet 131:1903–1924. https://doi.org/10.1007/s00122-018-3122-6
Li S, Wang L, Meng Y, Hao Y, Xu H, Hao M, Lan S, Zhang Y, Lv L, Zhang K, Peng X, Lan C, Li X, Zhang Y (2021) Dissection of genetic basis underpinning kernel weight-related traits in common wheat. Plants 10:713
Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X (2016) TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 14:1269–1280. https://doi.org/10.1111/pbi.12492
Manly KF, Cudmore JRH, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932. https://doi.org/10.1007/s00335-001-1016-3
Manning KS, Cooper TA (2017) The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol 18:102–114. https://doi.org/10.1038/nrm.2016.139
Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Fdr D, Balliau T, Bt V, Mn D, Trs T-L, Quilleré I, Coque M, Gallais A, Ma-Ba G-M, Bethencourt L, Habash DZ, Lea PJ, Charcosset A, Perez P, Murigneux A, Sakakibara H, Edwards KJ, Hirel B (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18:3252–3274. https://doi.org/10.1105/tpc.106.042689
Mayr C (2019) What are 3’ UTRs doing? Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a034728
Mir R, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney R (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125:625–645. https://doi.org/10.1007/s00122-012-1904-9
Mohler V, Albrecht T, Castell A, Diethelm M, Schweizer G, Hartl L (2016) Considering causal genes in the genetic dissection of kernel traits in common wheat. Theor Appl Genet 57:467–476. https://doi.org/10.1007/s13353-016-0349-2
Morbitzer R, Römer P, Boch J, Lahaye T (2010) Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci USA 107:21617–21622. https://doi.org/10.1073/pnas.1013133107
Németh E, Nagy Z, Pécsváradi A (2018) Chloroplast glutamine synthetase, the key regulator of nitrogen metabolism in wheat, performs its role by fine regulation of enzyme activity via negative cooperativity of its subunits. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00191
Nigro D, Gadaleta A, Mangini G, Colasuonno P, Marcotuli I, Giancaspro A, Giove SL, Simeone R, Blanco A (2019) Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta 249:1157–1175. https://doi.org/10.1007/s00425-018-03075-1
Nigro D, Fortunato S, Giove SL, Mazzucotelli E, Gadaleta A (2020) Functional validation of glutamine synthetase and glutamate synthase genes in durum wheat near isogenic lines with QTL for high GPC. Int J Mol Sci. https://doi.org/10.3390/ijms21239253
Obara M, Kajiura M, Fukuta Y, Yano M, Hayashi M, Yamaya T, Sato T (2001) Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J Exp Bot 52:1209–1217. https://doi.org/10.1093/jexbot/52.359.1209
Oliveira IC, Coruzzi GM (1999) Carbon and Amino Acids Reciprocally Modulate the Expression of Glutamine Synthetase in Arabidopsis1. Plant Physiol 121:301–310. https://doi.org/10.1104/pp.121.1.301
Pang Y, Liu C, Wang D, St. Amand P, Bernardo A, Li W, He F, Li L, Wang L, Yuan X, Dong L, Su Y, Zhang H, Zhao M, Liang Y, Jia H, Shen X, Lu Y, Jiang H, Wu Y, Li A, Wang H, Kong L, Bai G, Liu S (2020) High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol Plant 13:1311–1327. https://doi.org/10.1016/j.molp.2020.07.008
Park CJ, Seo YS (2015) Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J 31:323–333. https://doi.org/10.5423/PPJ.RW.08.2015.0150
Rajaram S, Borlaug NE, van MG (2002) CIMMYT international wheat breeding. FOA Corporate Document Repository. http://www.fao.org/DOCREP/006/Y4011E/Y4011E00.HTM.
Rao SJ, Chatterjee S, Pal JK (2017) Untranslated regions of mRNA and their role in regulation of gene expression in protozoan parasites. J Biosci 42:189–207. https://doi.org/10.1007/s12038-016-9660-7
Ren T, Fan T, Chen S, Li C, Chen Y, Ou X, Jiang Q, Ren Z, Tan F, Luo P, Chen C, Li Z (2021) Utilization of a Wheat55K SNP array-derived high-density genetic map for high-resolution mapping of quantitative trait loci for important kernel-related traits in common wheat. Theor Appl Genet 134:807–821. https://doi.org/10.1007/s00122-020-03732-8
Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Shiferaw B, Smale M, Braun HJ, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur 5:291–317. https://doi.org/10.1007/s12571-013-0263-y
Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34:635–645. https://doi.org/10.1046/j.1365-313x.2003.01753.x
Swarbreck SM, Defoin-Platel M, Hindle M, Saqi M, Habash DZ (2011) New perspectives on glutamine synthetase in grasses. J Exp Bot 62:1511–1522. https://doi.org/10.1093/jxb/erq356
Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, Yamaya T (2005) Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J 42:641–651. https://doi.org/10.1111/j.1365-313X.2005.02406.x
Taira M, Valtersson U, Burkhardt B, Ludwig RA (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16:2048–2058. https://doi.org/10.1105/tpc.104.022046
Thomsen HC, Eriksson D, Møller IS, Schjoerring JK (2014) Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci 19:656–663. https://doi.org/10.1016/j.tplants.2014.06.002
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539. https://doi.org/10.1105/tpc.5.11.1529
Urriola J, Rathore KS (2015) Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res 24:397–407. https://doi.org/10.1007/s11248-014-9852-6
Wang Y, Teng W, Wang Y, Ouyang X, Xue H, Zhao X, Gao C, Tong Y (2020) The wheat cytosolic glutamine synthetase GS1.1 modulates N assimilation and spike development by characterizing CRISPR-edited mutants. bioRxiv:2020.2009.2003.281014. doi:https://doi.org/10.1101/2020.09.03.281014
Wen S, Zhang M, Tu K, Fan C, Tian S, Bi C, Chen Z, Zhao H, Wei C, Shi X, Yu J, Sun Q, You M (2022) A major quantitative trait loci cluster controlling three components of yield and plant height identified on chromosome 4B of common wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2021.799520
Xiong H, Li Y, Guo H, Xie Y, Zhao L, Gu J, Zhao S, Ding Y, Liu L (2021) Genetic Mapping by integration of 55K SNP array and KASP markers reveals candidate genes for important agronomic traits in hexaploid wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2021.628478
Xu D, Wen W, Fu L, Li F, Li J, Xie L, Xia X, Ni Z, He Z, Cao S (2019) Genetic dissection of a major QTL for kernel weight spanning the Rht-B1 locus in bread wheat. Theor Appl Genet. https://doi.org/10.1007/s00122-019-03418-w
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21:281–288. https://doi.org/10.1046/j.1365-313x.2000.00685.x
Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7:555–560. https://doi.org/10.1016/S1360-1385(02)02362-2
Yang L, Zhao D, Meng Z, Xu K, Yan J, Xia X, Cao S, Tian Y, He Z, Zhang Y (2020) QTL mapping for grain yield-related traits in bread wheat via SNP-based selective genotyping. Theor Appl Genet 133:857–872. https://doi.org/10.1007/s00122-019-03511-0
Yin H, Yang F, He X, Du X, Mu P, Ma W (2022) Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response. Crop J. https://doi.org/10.1016/j.cj.2022.01.003
Yu Z, She M, Zheng T, Diepeveen D, Islam S, Zhao Y, Zhang Y, Tang G, Zhang Y, Zhang J, Blanchard CL, Ma W (2021) Impact and mechanism of sulphur-deficiency on modern wheat farming nitrogen-related sustainability and gliadin content. Commun Biol 4:945. https://doi.org/10.1038/s42003-021-02458-7
Zhang J, Huang S, Fosu-Nyarko J, Dell B, McNeil M, Waters I, Moolhuijzen P, Conocono E, Appels R (2008) The genome structure of the 1-FEH genes in wheat (Triticum aestivum L.): new markers to track stem carbohydrates and grain filling QTLs in breeding. Mol Breed 22:339–351. https://doi.org/10.1007/s11032-008-9179-1
Zhang J, Dell B, Conocono E, Waters I, Setter T, Appels R (2009) Water deficits in wheat: fructosyl exohydrolase (1-FEH) mRNA expression and relationship to soluble carbohydrate concentrations in two varieties. New Phytol 181:843–850
Zhang J, Dell B, Biddulph B, Drake-Brockman F, Walker E, Khan N, Wong D, Hayden M, Appels R (2013) Wild-type alleles of Rht-B1 and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol Breed 32:771–783. https://doi.org/10.1007/s11032-013-9905-1
Zhang J, Dell B, Biddulph B, Khan N, Xu Y, Luo H, Appels R (2014a) Vernalization gene combination to maximize grain yield in bread wheat (Triticum aestivum L.) in diverse environments. Euphytica 198:439–454. https://doi.org/10.1007/s10681-014-1120-6
Zhang Y, Liu J, Xia X, He Z (2014b) TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed 34:1097–1107. https://doi.org/10.1007/s11032-014-0102-7
Zhang J, Xu Y, Chen W, Dell B, Vergauwen R, Biddulph B, Khan N, Luo H, Appels R, Van den Ende W (2015) A wheat 1-FEH w3 variant underlies enzyme activity for stem WSC remobilization to grain under drought. New Phytol 205:293–305. https://doi.org/10.1111/nph.13030
Zhang Z, Xiong S, Wei Y, Meng X, Wang X, Ma X (2017) The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci Rep 7:1000. https://doi.org/10.1038/s41598-017-01071-1
Zhang J, She M, Yang R, Jiang Y, Qin Y, Zhai S, Balotf S, Zhao Y, Anwar M, Alhabbar Z, Juhász A, Chen J, Liu H, Liu Q, Zheng T, Yang F, Rong J, Chen K, Lu M, Islam S, Ma W (2021) Yield-related QTL clusters and the potential candidate genes in two wheat DH populations. Int J Mol Sci 22:11934
Zhang J (2008) Water deficit in bread wheat: characterisation using genetic and physiological tools. PhD thesis, Murdoch University
Zhao Y (2019) Genetic dissection of wheat nitrogen use efficiency related traits. PhD Thesis, Murdoch University
Zhu X-F, Zhang H-P, Hu M-J, Wu Z-Y, Jiang H, Cao J-J, Xia X-C, Ma C-X, Chang C (2016) Cloning and characterization of Tabas1-B1 gene associated with flag leaf chlorophyll content and thousand-grain weight and development of a gene-specific marker in wheat. Mol Breed 36:142. https://doi.org/10.1007/s11032-016-0563-y
Acknowledgements
This work was supported by Murdoch University and the Australia Grains Research and Development Corporation (GRDC) (Grant Number UMU00048), the China Scholarship Council (CSC) (Grant Number CSC201906910028), the Department of Primary Industries and Regional Development (DPIRD), Western Australia. We thank Dr. Dean Diepeveen and his colleagues in DPIRD, and Dr. Hugo. Alsono-Cantabrana from the GRDC for their support and assistance. We thank InterGrain, Western Australia, for providing the Westonia and Kauz DH population.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
WM, JZ and SI conceived the project and designed the study; JZ, YZ, SI and WM carried out field experiments; FY, JZ and QL performed the gene sequencing, molecular marker development and data analysis; JZ, WY and WM provided the resources for the study; FY and JZ wrote the original draft of the manuscript; WM and SI provided extensive revision and editing; WM, SI and WY supervised and managed the project. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Susanne Dreisigacker.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, F., Zhang, J., Zhao, Y. et al. Wheat glutamine synthetase TaGSr-4B is a candidate gene for a QTL of thousand grain weight on chromosome 4B. Theor Appl Genet 135, 2369–2384 (2022). https://doi.org/10.1007/s00122-022-04118-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-022-04118-8