Abstract
Downy mildew, caused by Plasmopara halstedii, is one of the most destructive diseases in cultivated sunflower (Helianthus annuus L.). The dominant resistance locus Pl ARG originates from silverleaf sunflower (H. argophyllus Torrey and Gray) and confers resistance to all known races of P. halstedii. We mapped Pl ARG on linkage group (LG) 1 of (cms)HA342 × ARG1575-2, a population consisting of 2,145 F2 individuals. Further, we identified resistance gene candidates (RGCs) that cosegregated with Pl ARG as well as closely linked flanking markers. Markers from the target region were mapped with higher resolution in NDBLOSsel × KWS04, a population consisting of 2,780 F2 individuals that does not segregate for Pl ARG . A large-insert sunflower bacterial artificial chromosome (BAC) library was screened with overgo probes designed for markers RGC52 and RGC151, which cosegregated with Pl ARG . Two RGC-containing BAC contigs were anchored to the Pl ARG region on LG 1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sunflower (Helianthus annuus L.) is one of the major oilseed crops cultivated worldwide in 25 mio. ha (FAOSTAT 2008). Plasmopara halstedii (Farl.) Berl. & de Toni is a soil-, seed- and wind-borne pathogen causing downy mildew in sunflower, which can survive up to 10 years in the soil in the form of oospores (EPPO/CABI 1997). Downy mildew is a common sunflower disease responsible for significant yield loss and can be controlled by fungicides and cultivation of resistant hybrids. Several Pl loci (Pl 1 –Pl 13 , Pl ARG , Pl v –Pl z ) have been described, which confer resistance to one or more races of P. halstedii (Dußle et al. 2004; Miller and Gulya 1991; Molinero-Ruiz et al. 2002b, 2003; Mulpuri et al. 2009; Rahim et al. 2002; Vranceanu and Stoenescu 1970; Vranceanu et al. 1981; Zimmer and Kinman 1972). The Pl loci were discovered in wild H. annuus ecotypes, as in the case of Pl 6 (Miller and Gulya 1991), or were introduced from other wild Helianthus species. Pl 5 was introgressed from H. tuberosus (Vranceanu et al. 1981) and Pl 7 from H. praecox (Miller and Gulya 1991), while Pl 8 and Pl ARG both originated from H. argophyllus (Miller and Gulya 1991; Seiler et al. 1991). Two types of resistance are known: P. halstedii is limited to the roots in seedlings with type I resistance but in seedlings with type II resistance it grows through the hypocotyls and a slight sporulation can be observed on the hypocotyls (Mouzeyar et al. 1994). Sackston (1990) called the phenomenon observed in type II resistance “cotyledon limited infection” (CLI).
In the last decade, new races of P. halstedii were discovered in the cultivation areas of sunflower (Gulya et al. 1991; Molinero-Ruiz et al. 2002a; Tourvieille de Labrouhe et al. 2000). Delmotte et al. (2008) investigated 24 isolates collected in France between 1966 and 2006 and concluded that the biology of P. halstedii, the multiple introductions of the pathogen into France and the selection pressure caused by the use of host resistance genes may have caused the spread of new races of P. halstedii. Gulya (2007) reported the existence of at least 35 pathotypes. Several races developed tolerance to metalaxyl and mefenoxam, the only effective fungicides available (Albourie et al. 1998; Molinero-Ruiz et al. 2005; Molinero-Ruiz and Melero-Vara 2003; Spring et al. 2006). Therefore, investigation of the structure and functionality of Pl loci is necessary in order to exploit them effectively and durably in plant breeding.
Pl loci were initially considered single independent genes (Miller and Gulya 1991), but genetic mapping and segregation studies indicated that some loci consist of clusters of resistance genes with different specificities (Vear et al. 1997). Mouzeyar et al. (1995) mapped the first Pl locus (Pl 1 ) on group 1 of the CARTISOL map (Gentzbittel et al. 1995), which corresponds to LG 8 of the publicly available simple sequence repeat (SSR) map constructed by Tang et al. (2002). Roeckel-Drevet et al. (1996) and Vear et al. (1997) mapped Pl 2 and Pl 6 , respectively, and found that both loci are located in the same genomic region as Pl 1 . Similarly, it was shown that LG 13 carries Pl 5 and Pl 8 (Bert et al. 2001; Radwan et al. 2003). Dußle et al. (2004) showed that Pl ARG is located on LG 1 and is unlinked to all previously mapped Pl loci. Pl ARG was introgressed from H. argophyllus and mediates resistance to all known races of P. halstedii (Seiler et al. 1991, G. Seiler, personal communication). Recently, Mulpuri et al. (2009) mapped Pl 13 on LG 1, but it is unlinked to Pl ARG and has its position at the lower end of LG 1.
Many plant disease resistance (R) genes have been discovered and cloned in the recent years. Five major classes of R-genes are known, the largest of which is the NBS-LRR class, consisting of the TIR-NBS-LRR and non-TIR-NBS-LRR subclasses (Dangl and Jones 2001). Most of the known R-genes form tightly linked gene families (Hulbert et al. 2001).
Knowing that the resistance genes Pl 1 , Pl 2 , Pl 6 , Pl 7 on LG 8 (Mouzeyar et al. 1995; Roeckel-Drevet et al. 1996; Vear et al. 1997) and Pl 5 , Pl 8 on LG 13 (Bert et al. 2001; Radwan et al. 2003) are clustered, we may as well assume that Pl ARG might also coincide with a cluster of resistance genes. Our study aims at the fine mapping and map-based cloning of the Pl ARG locus. Our objectives were: (1) to identify markers flanking Pl ARG and find recombinants in the target region, (2) to carry out phenotypic analyses with different races of P. halstedii and elucidate the genetic structure of Pl ARG , (3) to define markers as starting points for a map-based cloning approach, (4) to select bacterial artificial chromosome (BACs) for sequencing to identify candidate genes for Pl ARG , and (5) to investigate whether Pl ARG is a single gene or a complex resistance gene cluster.
Materials and methods
Plant material
In the present study, we used five resistant inbred lines: ARG1575-2, RHA419, RHA420, RHA443 and 79ARGMTP. ARG1575-2 is a homozygous resistant inbred line that carries the Pl ARG locus and mediates resistance to all known races of P. halstedii (Seiler et al. 1991). To our knowledge, there is no report about virulent races of P. halstedii that overcome the Pl ARG resistance. ARG1575-2 was derived by crossing H. argophyllus accession 1575 (PI 468651) with cmsHA89 followed by two generations of backcrossing with cmsHA89 and five selfing generations (Seiler et al. 1991). The homozygous resistant inbred lines RHA419, RHA420 and RHA443 carry Pl ARG and are all derived from ARG1575-2. RHA419 and RHA420 were derived from the cross RHA373 × ARG1575-2 (Miller et al. 2002), whereas RHA443 is an F6:7 restorer line selected from the cross RHA426/RHA419//RHA377/AS4379 (Miller et al. 2006). 79ARGMTP carries a downy mildew resistance on LG 1, which is likely to be Pl ARG , and was developed from the cross of H. argophyllus MPHE-92 with H. annuus FS20-6-2 at INRA, Montpellier (Vear et al. 2008a).
Three susceptible inbred lines, HA342, NDBLOSsel and KWS04 were used in this study. The susceptible inbred line HA342 was available both in the normal as well as in the PET1 cytoplasmic male sterility (CMS) cytoplasm. HA342 is derived from a single BC1F4 plant from the cross HA89*2/Pervenets and has high oleic seed content (Miller et al. 1987). NDBLOSsel is an inbred line selected from the germplasm pool ND-BLOS (Roath et al. 1987) and was used for QTL mapping of Sclerotinia midstalk rot resistance (Micic et al. 2005). KWS04 is proprietary line of KWS SAAT AG (Einbeck, Germany) and is used for the development of hybrid varieties.
Development of mapping populations
To fine map the target Pl ARG region on LG 1, we developed a segregating population from the cross of (cms)HA342 and ARG1575-2, which comprised of two subpopulations with a total of 2,145 F2 individuals. The subpopulation cmsHA342 × ARG1575-2 consisted of 1,065 F2 individuals, whereas the subpopulation HA342 × ARG1575-2 consisted of 1,080 F2 individuals. In this paper (cms)HA342 × ARG1575-2 refers to the whole population of 2,145 F2 individuals. We also developed population NDBLOSsel × KWS04 comprising of 2,780 F2 individuals to increase the mapping resolution of the targeted genomic region. NDBLOSsel and KWS04 are both highly polymorphic lines and susceptible to P. halstedii; therefore NDBLOSsel × KWS04 did not segregate for Pl ARG . DNA was isolated from dried leaves of each F2 plant and parental lines using the CTAB extraction method (Doyle and Doyle 1990).
Resistance tests
Initially, we evaluated resistance to downy mildew in a subset of 183 F2:3 families of the subpopulation cmsHA342 × ARG1575-2 using the whole seedling immersion test (Gulya 1996) with a suspension of P. halstedii race 730 at a concentration of 40,000 spores/ml. Next, we evaluated only informative lines which were selected as described below. The resistance of F2 plants was investigated by testing 16–40 F3 seedlings per F2 individual. Seedlings were considered susceptible when high fungal sporulation was evident on the cotyledons and resistant when no sporulation or only spurious sporulation was observed on the cotyledons. Because of the occurrence of CLI in plants which carry the resistance gene Pl ARG , progenies with ambiguous phenotype were re-tested using 2–10 F3:4 families. F2 plants were classified as homozygous susceptible, homozygous resistant or heterozygous according to the phenotypes of F3 or F4 families and the goodness-of-fit of observed segregation ratios was tested.
Linkage mapping analyses
We developed an anchor map of LG 1 for 475 F2 individuals of cmsHA342 × ARG1575-2 using seven polymorphic codominant SSR markers of the public sunflower linkage map (Tang et al. 2003; Tang et al. 2002), which were identified by Dußle et al. (2004). To increase the density of makers in the target Pl ARG region, 22 additional markers were screened for polymorphism among (cms)HA342, ARG1575-2, NDBLOSsel and KWS04. Thirteen SSR markers (Tang et al. 2002, 2003; Yu et al. 2003), six single nucleotide polymorphism (SNP) markers (Lai et al. 2005a) and three resistance gene candidates (RGCs) (Radwan et al. 2008) were analyzed using a LI-COR DNA-Analyzer 4300 (LI-COR Biosciences, Bad Homburg, Germany) or the single strand conformation polymorphism (SSCP) method (Slabaugh et al. 1997). HT211 and RGC151 were converted into cleaved amplified polymorphic sequence (CAPS) markers and were resolved with 3.0% agarose gels after digesting with TaqI and RsaI, respectively.
Overall, 19 SSR, SNP, CAPS and RGC markers were used for linkage analysis of (cms)HA342 × ARG1575-2, and 10 SSR, SNP and CAPS markers were used to construct the linkage map of NDBLOSsel × KWS04. To investigate the portion of H. argophyllus genome in the resistant inbred line ARG1575-2 marker scores of ARG1575-2 and cmsHA89 were compared with 14 codominant SSR and two CAPS loci of LG 1, and 94 randomly distributed SSR markers on the rest of the genome (LG 2–LG 17). We also compared the haplotypes of the resistant lines ARG1575-2, RHA419, RHA420, RHA443 and 79ARGMTP to estimate the size of the introgressed segment of H. argophyllus using the same set of 14 codominant SSR and two CAPS markers of LG 1.
The goodness-of-fit test was performed for the H o hypothesis to observe a segregation ratio of 1:2:1 for all polymorphic markers under study in the mapping populations. Maps for the populations (cms)HA342 × ARG1575-2 and NDBLOSsel × KWS04 were constructed with JOINMAP 4.0 (Van Ooijen 2006) using a LOD threshold of >3.0 and the Kosambi mapping function (Kosambi 1944).
Selecting informative lines carrying recombination events in the target region
We used the strategy outlined in Fig. 1 to identify recombination events in the region of Pl ARG . Only recombinant lines were phenotypically tested for fine mapping. Briefly, 2,145 F2 individuals of (cms)HA342 × ARG1575-2 were genotyped with ORS610 and ORS371, which flank Pl ARG , and with ORS662, which cosegregated with Pl ARG . 188 recombinant F2 individuals were analyzed with 16 additional polymorphic markers to increase the marker density in the target region, while F2:3 or F2:4 progenies of 108 recombinant F2 individuals were subsequently phenotyped to confirm the genomic location of Pl ARG . Next, ORS509, HT244 and HT446 were used to narrow down the interval harboring Pl ARG . We evaluated 25 homozygous F4 seedlings for each F2 individual that had a recombination event in the interval of ORS509, HT244 and HT446 for resistance to P. halstedii races 330, 100, 710 and 730 (Fig. 1).
High resolution mapping strategy for the identification of informative recombinant lines in cross (cms)HA342 × ARG1575-2. The F2 population consisted of 2,145 lines and was screened in two steps. First, the whole population was screened with markers ORS610, ORS662 and ORS371 and 188 recombinant lines were selected and, subsequently, genotyped and phenotyped. Second, lines with recombination events between the closely linked flanking markers ORS509, HT244 and HT446 were selfed and homozygous recombinant lines were tested with the four P. halstedii races 730, 100, 330, and 710
BAC library screening and mapping of BAC end sequences
Overgo probes (Han et al. 2000; Zheng et al. 2006) were designed from the sequence of RGC52 and RGC151, which cosegregated with Pl ARG . The probes were used for high-density filter hybridization to the 8× genome coverage large-insert sunflower BAC library, HA_HBa, constructed from HA383 and publicly available through the Clemson University Genomics Institute (CUGI, http://www.genome.clemson.edu). Positive BAC clones were confirmed by colony PCR and plasmids were isolated using the alkaline lysis method (Birnboim and Doly 1979). Plasmid DNA was used for high information content fingerprinting (HICF) (Ding et al. 1999; Luo et al. 2003) and BAC end sequencing (BES). BAC contigs were assembled using the FPC software v.8.5.3 (Soderlund et al. 1997). Inserts of positive BAC clones were sequenced from both ends using the T7 and M13 sequencing primers. Primers were designed from BAC end sequences with Primer3 (Rozen and Skaletsky 2000). To map the assembled BAC contigs in (cms)HA342 × ARG1575-2 and NDBLOSsel × KWS04, the polymorphic markers Co3-4_T7, Co3-2_T7 and Co7-11_M13 were analyzed which were designed from BES P202H01_T7, BES P0323D15_T7 and BES P399D04_M13 (Supplementary Table 1). PCR was carried out in a final volume of 10 μl using 275 nM of each primer (Table 1). In the presence of 1× Taq polymerase buffer, 2 mM MgCl2, 0.2 mM of each dNTP and 0.4 U Taq polymerase (Q-Biogene, Illkirch, Germany) PCR was performed under the following conditions: intitial denaturation at 95°C for 5 min, 35 PCR cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 1:15 min and an extension at 72°C for 10 min. The PCR products of Co3-4_T7 and Co7-11_M13 were analyzed on 1.5% agarose gels and products of Co3-2_T7 were analyzed on a SSCP gel.
Results
Localization of Pl ARG on LG 1
An anchor linkage map of cmsHA342 × ARG1575-2 was constructed using seven polymorphic SSR markers. ORS1182, ORS610, ORS1128 and ORS543 cosegregated at the upper end of LG 1, whereas ORS662, ORS959 and ORS371 mapped 0.5, 3.6 and 5.2 cM from the upper end, respectively. F2:3 or F2:4 families of 183 F2 individuals that produced sufficient seeds were evaluated for resistance to downy mildew. We observed 25 homozygous resistant, 114 segregating, and 44 homozygous susceptible progenies, which differs significantly from the expected 1:2:1 Mendelian segregation ratio of Pl ARG (X 2 = 15.01, DF = 2, p ≤ 0.0006). Pl ARG cosegregated with ORS662 (map not shown).
Fine mapping of the target region
Population (cms)HA342 × ARG1575-2
To increase the genetic resolution around Pl ARG we genotyped 2,145 F2 individuals of (cms)HA342 × ARG1575-2 as described in Fig. 1. ORS610 and ORS371 mapped at the upper and lower end of LG 1, respectively, and flank Pl ARG , which cosegregates with ORS662. The three markers were used to screen 2,145 F2 individuals. All markers showed significant segregation distortion with a lack of the ARG1575-2 allele in the subpopulation cmsHA342 × ARG1575-2, but did not deviate from the expected 1:2:1 segregation ratio in HA342 × ARG1575-2 (Table 2).
In total, we identified 188 F2 individuals that carried a recombination event between the two flanking markers ORS610 and ORS371. The F2 individuals were phenotyped and genotyped with 16 polymorphic markers between ORS610 and ORS371. To confirm the genomic position of Pl ARG , we tested F2:3 or F2:4 progenies of 108 out of the 188 recombinant F2 individuals (Fig. 2a). The remaining 80 recombinant F2 individuals could not be tested due to lack of sufficient seed especially for cmsHA342 × ARG1575-2 (Supplementary Fig. 1).
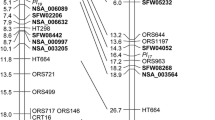
a LG 1 of (cms)HA342 × ARG1575-2 constructed with 2,145 F2 individuals. b Partial LG 1 map of NDBLOSsel × KWS04 constructed with 528 F2 individuals. c Partial LG 1 map of NDBLOSsel × KWS04 constructed with 2,780 F2 individuals. Markers with an asterisk were screened in all individuals, while all other markers were screened in recombinant lines only. Pl ARG is shown in bold. Maps are not drawn to scale and distances on the left side of the map correspond to centiMorgan
The genetic map of LG 1 in (cms)HA342 × ARG1575-2 spanned 4.2 cM (Fig. 2a). ORS610, ORS543, ORS1128, CRT272, and ORS1182 cosegregated at the upper end of LG 1, whereas Pl ARG cosegregated with ORS662, HT211, RGC52a, RGC52b, RGC151, HT722, and ORS716 0.3 cM from the upper end. ORS509, HT244 on one side and HT446 on the other side flanked Pl ARG at a distance of 0.2 cM and 0.1 cM, respectively. More recombination events were observed below Pl ARG and ORS053, ORS959 and ORS371 mapped 1.9, 2.3, and 3.9 cM from Pl ARG , respectively. Thus, we identified closely linked markers on both sides of Pl ARG to use for fine mapping of the Pl ARG genomic region.
Of the 188 F2 individuals selected initially (Fig. 1), only 15 F2 individuals were selected in the second step and revealed a recombination event between the closely linked flanking markers ORS509, HT244 and HT446. Five of the 15 recombinant lines produced sufficient seed to develop homozygous recombinant progenies. Four homozygous lines were resistant to P. halstedii race 730, and one homozygous line was susceptible to P. halstedii race 730 and showed the same pattern of resistance after infection with P. halstedii races 100, 330, 710.
Population NDBLOSsel × KWS04
Suppressed recombination was evident in (cms)HA342 × ARG1575-2 comparing with the linkage map of Tang et al. (2002, 2003), which is likely due to the introgression of Pl ARG . To obtain a higher mapping resolution in the target region, we developed the intraspecific population NDBLOSsel × KWS04 of 2,780 F2 individuals that did not carry an introgression from Pl ARG . Initially, 528 F2 individuals were screened with ORS610, ORS662 and ORS053 from LG 1. For 198 F2 individuals a recombination event was detected between ORS610 and ORS053. These individuals were screened with four additional polymorphic SSR markers and resulted in a map based on 528 F2 individuals (Fig. 2b) that spanned 21.2 cM between ORS610 and ORS053. ORS610, ORS1128, CRT272, and ORS1182 were mapped within 12.5 cM. ORS1182 is the closest flanking marker above Pl ARG . Hence, the target interval (ORS1182–HT446) spans 2.4 cM in NDBLOSsel × KWS04.
Additional 2,252 F2 individuals of NDBLOSsel × KWS04 were screened with ORS1182 and HT446 to identify recombinants in this high-resolution mapping population. Altogether, 99 recombinant F2 individuals were identified in the target region and were screened with HT244, HT211, ORS662 and ORS675. HT211 and ORS662 which cosegregated with Pl ARG in (cms)HA342 × ARG1575-2 were mapped with higher resolution in NDBLOSsel × KWS04 0.2 cM apart (Fig. 2c).
Screening the large-insert sunflower BAC library HA383 and mapping BAC end sequences
Screening the HA383 BAC library with overgo probes designed for RGC52 and RGC151 identified 18 positive BAC clones for RGC52 and 9 positive BAC clones for RGC151. FPC assembles of the 18 BAC clones of RGC52 resulted in two contigs: 11 BACs in contig 2 and 7 BACs in contig 7. BAC clones of RGC151 were assembled all together in contig 3. BES generated 19, 17 and 14 BAC end sequences for contigs 2, 3 and 7. After quality control and vector sequence trimming, the BAC end sequence lengths ranged from 149 to 834 bp. Contig 2 was assigned to LG 16 (W. Gao, unpublished results). Therefore, primers were designed on the BAC end sequences of contig 3 and contig 7 (Supplementary Material Table 1) and amplicons were tested for polymorphism between (cms)HA342 and ARG1575-2 and between NDBLOSsel and KWS04. Marker Co3-4_T7 from contig 3 was polymorphic between (cms)HA342 and ARG1575-2 and marker Co3-2_T7 was polymorphic between NDBLOSsel × KWS04. Marker Co7-11_M13 from contig 7 was polymorphic for both mapping populations. Overall, the 188 recombinant F2 individuals (ORS610/ORS371) of (cms)HA342 × ARG1575-2 were analyzed with markers Co3-4_T7, and Co7-11_M13 and the 99 recombinant F2 individuals (ORS1182/HT446) of NDBLOSsel × KWS04 were genotyped with markers Co3-2_T7 and Co7-11_M13. Map calculation for the final maps was done for the whole population (cms)HA342 × ARG1575-2 with 2,145 F2 individuals and NDBLOSsel × KWS04 with 2,780 F2 individuals. Co3-4_T7 and Co7-11_M13 and consequently contig 3 and contig 7 cosegregated with Pl ARG in (cms)HA342 × ARG1575-2 (Fig. 2a). HT211 and ORS662 flanked contig 3 and contig 7 on LG 1 in NDBLOSsel × KWS04 (Fig. 2c).
Origin of LG 1 in the resistant line ARG1575-2
To explain the suppressed recombination of (cms)HA342 × ARG1575-2, we compared the alleles of ARG1575-2 and cmsHA89 for 14 SSR and two CAPS markers of LG 1. The resistant inbred line ARG1575-2 is expected to carry only 12.5% of H. argophyllus genome after two backcrosses with cmsHA89. We found that ARG1575-2 and cmsHA89 carry different alleles in all markers tested on LG 1 with the exception of HT324 (Table 3). We could not investigate whether the HT324 allele of ARG1575-2 originated from H. argophyllus or from cmsHA89 because accession 1575 is no longer available. Analysis of 94 SSR markers randomly distributed on LG 2 to 17 revealed that 12.7% of them had shared alleles between ARG1575-2 and cmsHA89. To estimate the size of the introduced segment in other resistant inbred lines carrying Pl ARG we compared the marker alleles of ARG1575-2 and six inbred lines. The haplotypes of RHA419, RHA443 and RHA420 originate mostly from ARG1575-2 (Table 3). A recombination event occurred in RHA419 and RHA443 below Pl ARG in the interval of ORS053 and ORS959. Comparison of 79ARGMTP, which probably also carries Pl ARG , and FS20-6-2 showed no evidence of recombination on LG 1 (Table 3).
Discussion
Wild species are often used to broaden the genetic background of cultivated crops to increase yield (Singh and Ocampo 1997), oil content and quality (Seiler 2007), tolerance to abiotic stress (Miller and Seiler 2003), herbicide tolerance (Al-Khatiba et al. 1998) or to introduce male sterility alleles for developing hybrids (Leclercq 1969). However, wild species are most commonly used as donors of disease resistance genes (Jan et al. 2002, 2004a, b; Kuhl et al. 2001; Ling et al. 2004). The dominant monogenic locus Pl ARG originated from the wild species H. argophyllus and is an outstanding source of resistance, because of broad-spectrum resistance against all known races of P. halstedii.
Monogenic resistance genes are often not durable. Parlevliet (2002) reported that the presence of many major resistance genes and the occurrence of hypersensitive response, both of which are typical for Pl loci, are characteristics of non-durable resistance. Many major dominant genes for resistance to downy mildew have been described (Pl 1 –Pl 13 ) and Radwan et al. (2005) showed that the resistance mechanism of Pl 8 is associated with a hypersensitive response in the hypocotyls. Pl 8 confers till now resistance against all known races of downy mildew, but Pl 6 is an example where the resistance has been overcome by races 304 and 314 (Vear et al. 2007). A few studies have commented on extending the durability of Pl loci. Vear et al. (2008b) and Tourvieille de Labrouhe et al. (2008) suggested a combination of monogenic Pl loci with quantitative resistance against downy mildew. McDonald and Linde (2002) recommend pyramiding major resistance genes in one variety or growing cultivar mixtures containing genotypes with different major resistance genes. To realize these strategies molecular markers and knowledge of the genetic and functional basis of resistance are required.
Dußle et al. (2004) mapped Pl ARG in the telomeric region of LG 1 using 126 F2 individuals of cmsHA342 × ARG1575-2. In this study we successfully remapped Pl ARG and defined a new position on LG 1, because a larger mapping population of (cms)HA342 × ARG1575-2 was available and more phenotypic and genotypic data were collected. We enriched the target Pl ARG region with the closely linked flanking markers ORS509, HT244 and HT446 and with the newly identified cosegregating SSR markers ORS716, HT722 and SNP marker HT211 as well as the resistance gene candidates RGC151, RGC52a and RGC52b. The RGCs belong to the NBS-LRR class, but sequence information is not currently available to classify them in the TIR or non-TIR subclasses (Radwan et al. 2008). Here, we present the fine mapping of Pl ARG that will lay the groundwork for map-based cloning of this resistance locus.
Suppressed recombination on LG 1 in cross (cms)HA342 × ARG1575-2
ORS610, ORS1128, CRT272, and ORS1182 cosegregated at the upper end of LG 1 in (cms)HA342 × ARG1575-2. However in NDBLOSsel × KWS04 these markers span a genetic distance of 12.5 cM. Comparison of the marker interval ORS610–ORS053 between the two linkage maps showed that we were able to increase mapping resolution by a factor of ten in NDBLOSsel × KWS04. The same interval spanned 31.5 cM in the publicly sunflower SSR map of LG 1 published by Tang et al. (2003) confirming suppression of recombination in (cms)HA342 × ARG1575-2.
LG 1 of (cms)HA342 × ARG1575-2 can be regarded as an interspecific cross between silverleaf and cultivated sunflower, because no recombination occurred during the cross of ARG1575-2 with cmsHA89 or during the two backcross steps with cmsHA89. During the development of ARG1575-2, resistance to downy mildew was not tested before the BC2F5 generation (Seiler et al. 1991). Therefore, the retention of H. argophyllus alleles on LG 1 in ARG1575-2 is surprising since the remaining linkage groups (LG 2–17) contain a mixture of H. annuus and H. argophyllus alleles. This raises the question whether the inheritance of the whole LG 1 occurred randomly or not during the development of ARG1575-2. Other crosses from the original population did not contain Pl ARG , indicating that the discovery of this locus was probably random and at a very low frequency (G. Seiler, personal communication).
Suppressed recombination is often observed in populations that carry wild genome introgressions as in case of Mla introduced into cultivated barley from H. spontaneum (Wei et al. 1999), Run1, the grapevine powdery mildew resistance gene introduced from Muscadinia rotundifolia into Vitis vinifera (Barker et al. 2005), and Mi and Tm2-a, both introduced from Lycopersicon peruvianum into L. esculentum (Ganal et al. 1989; Kaloshian et al. 1998; Messeguer et al. 1991; Seah et al. 2004). Seah et al. (2004) showed that both resistant and susceptible lines carry two Mi-1 homolog clusters which are separated in the resistant genotypes by 300 Kb while in susceptible genotypes they are separated by a genomic fragment of unknown size. Molecular markers that flanked the resistant and susceptible loci had the same orientation, but markers in between the two clusters had an inverse orientation. Therefore, it was concluded that suppressed recombination may be due to chromosomal inversion. Comparing sequences near the Mi-1-homologs between susceptible and resistant genotypes showed blocks of homology, but also regions that have undergone considerable rearrangements (Seah et al. 2007).
In sunflower, comparative genetic linkage maps of H. annuus, H. petiolaris, H. anomalus, H. deserticola and H. paradoxus were established to study karyotypic evolution and a high rate of chromosomal rearrangements was observed (Burke et al. 2004; Lai et al. 2005b). Heesacker et al. (2009) studied comparative mapping between H. annuus and H. argophyllus and identified 10 collinear chromosomes, 9 chromosomal rearrangements, 3 putative segmental duplications and 2 putative whole chromosome duplications. LG 1 was collinear between H. annuus and H. argophyllus; therefore, chromosomal rearrangements are not likely the reason for suppressed recombination, but reduced homology may be a plausible explanation. Suppressed recombination was previously described in sunflower mapping populations of interspecific origin (Burke et al. 2002; Heesacker et al. 2009). However, in an intraspecific mapping population of H. argophyllus recombination was also suppressed near Pl ARG so that the reduced homology between silverleaf and cultivated sunflower seems not to be the sole cause of suppressed recombination (Heesacker et al. 2009).
Does the introgressed region influence fertility and segregation distribution in mapping populations?
Eighty-one percent of the cmsHA342 × ARG1575-2 F2 individuals showed a limited seed set producing less than 10 g of sunflower seed. ARG1575-2 must carry a restorer gene for the PET1 cytoplasm otherwise no seed production would be possible in the progenies. For 25% of the F2 individuals no seed set was observed. This fits the expected 3:1 segregation of a single dominant restorer gene. Since the backcrossing parent cmsHA89 possesses no restorer gene for the PET1 cytoplasm, the restorer gene must originate from H. argophyllus. Compared to the seed production of subpopulation HA342 × ARG1575-2 the seed set in subpopulation cmsHA342 × ARG1575-2 is reduced. The lack of plants homozygous for the ARG1575-2 allele in cmsHA342 × ARG1575-2 could be due to incomplete restoration of the PET1 cytoplasm based on unfavorable effects of the restorer or other genes that prohibit full male sterility. The restorer gene is not closely linked to Pl ARG , because only 64 of 209 F2 individuals which carry the alleles of ARG1575-2 in the target region had no seed production (data not shown). Abratti et al. (2008) described a monogenic restorer gene for the PET1 cytoplasm originating from H. argophyllus. They mapped the Rf3 gene on LG 7 using the population RHA340 × ZENB8. It was not analyzed if Rf3 is the restorer gene in ARG1575-2. Another possible cause for the reduced seed set could be the introgressed genomic segment from H. argophyllus since reduced seed set has been frequently observed in progenies of interspecific crosses. Chetelat and Meglic (2000) reported a reduced seed set of tomato back-cross lines that carry introgressions from Solanum lycopersicoides. Lai et al. (2005b) reported fertility reductions observed in hybrids derived from interspecific crosses in the genus Helianthus. Fertility problems in cmsHA342 × ARG1575-2 were also reflected by the significant distortion from the 1:2:1 segregation at loci in the target region. Distorted segregation was not observed in subpopulation HA342 × ARG1575-2, which also had a better seed set, thus, the introgression from H. argophyllus itself does not seem to severely influence fertility and segregation ratios in the target region in crosses with the normal cytoplasm.
Fine mapping of the target region Pl ARG
A prerequisite for efficient fine mapping of a given genomic region is the availability of closely linked flanking markers. We identified the closely linked flanking markers (ORS509, HT244 and HT446) at a distance of 0.2 cM above and 0.1 cM below Pl ARG . We identified F2 individuals which carry a recombination event between the markers to reduce phenotyping work with different races of P. halstedii only to genetically informative lines (Bauer and Graner 1995). Ultimately, five recombinant lines were screened with four different races of P. halstedii, but no differences in segregation patterns were observed. This may be due to the lack of recombination in the resistance gene cluster or due to extreme physical proximity of clustered resistance genes. In the study of Mi-1 locus discussed previously, Seah et al. (2007) concluded that lethality of recombination events within the inverted genomic region between the two clusters of Mi homologs might explain the observed linkage map contraction. Moreover, another explanation could be that Pl ARG is a single resistance gene mediating resistance against several races of P. halstedii, similar to RPP13, a single resistance gene that protects Arabidopsis thaliana against different isolates of the biotrophic oomycete Pernospora parasitica (now known as Hyaloperonospora arabidopsis) (Bittner-Eddy et al. 2000). Pl ARG may differ from the previously analyzed Pl loci, which clustered on LG 8 (Pl 1 , Pl 2 , Pl 6 and Pl 7 ) (Mouzeyar et al. 1995; Roeckel-Drevet et al. 1996; Vear et al. 1997) and LG 13 (Pl 5 and Pl 8 ) (Bert et al. 2002; Radwan et al. 2003). Further study is necessary to find out whether Pl ARG is a single resistance gene or a complex locus of genes and characterize the molecular structure of Pl ARG . One plausible approach is the map-based cloning of Pl ARG to solve the question about the structure and to answer to which class of resistance genes Pl ARG belongs.
Radwan et al. (2008) showed that RGCs are landmarks for the identification and isolation of R-genes in sunflower. They identified RGCs linked to the downy mildew resistance loci Pl ARG and Pl 8 , to the black rust resistance genes R 1 and R ADV , and to Or 5 , which protects sunflower against the parasitic weed Orobanche cumana Wallr. race E. Recently, Pl 13 was mapped on the lower end of LG 1 (Mulpuri et al. 2009) where Radwan et al. (2008) mapped several RGCs. We used RGCs cosegregating with Pl ARG to screen the large-insert sunflower HA383 BAC library. Two BAC contigs were identified in the target Pl ARG region on LG 1 with overgo probes of RGC52 and RGC151, which indicates that there is a cluster of RGCs in the target region and Pl ARG may be a part of it. A third contig identified with RGC52 mapped on LG 16 (W. Gao, unpublished results). A similar approach was used by Meyers et al. (1998) who screened a BAC library of lettuce cultivar Diana with a probe designed in the NBS region of RGC2, which cosegregated with the downy mildew resistance gene Dm3, and identified several BAC clones. The positive BAC clones revealed 22 distinct members of a resistance gene family and RGC2B was identified as a candidate gene by screening deletion mutants.
NDBLOSsel × KWS04 is a suitable population for fine-mapping BAC end sequences in the target region at a higher resolution compared to (cms)HA342 × ARG1575-2. The aim of this work is to select and sequence BACs that map in the target region using next-generation sequencing technologies.
References
Abratti G, Bazzalo ME, León A (2008) Mapping a novel fertility restoration gene in sunflower. In: Velasco I (ed) Proceedings of 17th international sunflower conference, Córdoba, Spain, pp 617–621
Albourie J-M, Tourvieille J, Tourvieille de Labrouhe D (1998) Resistance to metalaxyl in isolates of the sunflower pathogen Plasmopara halstedii. Eur J Plant Pathol 104:235–242
Al-Khatiba K, Baumgartner JR, Peterson DE, Currie RS (1998) Imazethapyr resistance in common sunflower (Helianthus annuus). Weed Sci 46:403–407
Barker CL, Donald T, Pauquet J, Ratnaparkhe MB, Bouquet A, Adam-Blondon AF, Thomas MR, Dry I (2005) Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor Appl Genet 111:370–377
Bauer E, Graner A (1995) Basic and applied aspects of the genetic analysis of the ym4 virus resistance locus in barley. Agronomie 15:469–473
Bert PF, Tourvielle de Labrouhe D, Philippon J, Mouzeyar S, Jouan I, Nicolas P, Vear F (2001) Identification of second linkage group carrying genes controlling resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor Appl Genet 103:992–997
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucl Acids Res 7:1513–1523
Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21:177–188
Burke JM, Tang S, Knapp SJ, Rieseberg LH (2002) Genetic analysis of sunflower domestication. Genetics 161:1257–1267
Burke JM, Lai Z, Salmaso M, Nakazato T, Tang S, Heesacker A, Knapp SJ, Rieseberg LH (2004) Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167:449–457
Chetelat RT, Meglic V (2000) Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor Appl Genet 100:232–241
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Delmotte F, Giresse X, Richard-Cervera S, M’Baya J, Vear F, Tourvieille J, Walser P, Tourvieille de Labrouhe D (2008) Single nucleotide polymorphisms reveal multiple introductions into France of Plasmopara halstedii, the plant pathogen causing sunflower downy mildew. Infect Genet Evol 8:534–540
Ding Y, Johnson MD, Colayco R, Chen YJ, Melnyk J, Schmitt H, Shizuya H (1999) Contig assembly of bacterial artificial chromosome clones through multiplexed fluorescence-labeled fingerprinting. Genomics 56:237–246
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dußle CM, Hahn V, Knapp SJ, Bauer E (2004) Pl Arg from Helianthus argophyllus is unlinked to other known downy mildew resistance genes in sunflower. Theor Appl Genet 109:1083–1086
EPPO/CABI (1997) Plasmopara halstedii. In: Smith I, McNamara D, Scott P, Holderness M (eds) Quarantine pests for Europe, vol 2. CABI International, Wallingford, p 1425
Ganal MW, Young ND, Tanksley SD (1989) Pulsed field gel electrophoresis and physical mapping of large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol Gen Genet 215:395–400
Gentzbittel L, Vear F, Zhang YX, Bervillé A, Nicolas P (1995) Development of a consensus linkage RFLP map of cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 90:1079–1086
Gulya TJ (1996) Everything you should know about downy mildew testing but were afraid to ask. In: Proceedings of the 18th sunflower research workshop, January 11–12, 1996, Fargo, ND, USA, pp 39–48
Gulya TJ (2007) Distribution of Plasmopara halstedii races from sunflower around the world. In: Lebeda A, Spencer-Phillips PTN (eds) Advances in downy mildew research proceedings of 2nd international downy mildew symposium, Olomouc, Czech Republic, Palacky University and JOLA, Kastelec na Hané, Czech Republic, pp 121–134
Gulya TJ, Sacksston WE, Virany F, Masierevic S, Rashid KY (1991) New races of the sunflower downy mildew pathogen (Plasmopara halstedii) in Europe and North and South America. J Phytopathol 132:303–311
Han CS, Sutherland RD, Jewett PB, Campbell ML, Meincke LJ, Tesmer JG, Mundt MO, Fawcett JJ, Kim U-J, Deaven LL, Doggett NA (2000) Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res 10:714–721
Heesacker AF, Bachlava E, Brunick RL, Burke JM, Rieseberg LH, Knapp SJ (2009) Comparative mapping identifies multiple chromosomal rearrangements and duplications in the common and silverleaf sunflower genomes. Plant Genome 2:1–14
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Jan CC, Fernández-Martínez JM, Ruso J, Munoz-Ruz J (2002) Registration of four sunflower germplasms with resistance to Orobanche cumana race F. Crop Sci 42:2217–2218
Jan CC, Quresh Z, Gulya TJ (2004a) Registration of seven rust resistant sunflower germplasms. Crop Sci 44:1887–1888
Jan CC, Tan AS, Gulya TJ (2004b) Registration of four downy mildew resistant sunflower germplasms. Crop Sci 44:1887
Kaloshian I, Yaghoobi J, Liharska T, Hontelez J, Hanson D, Hogan P, Jesse T, Wijbrandi J, Simons G, Vos P, Zabel P, Williamson VM (1998) Genetic and physical localization of the root-knot nematode resistance locus Mi in tomato. Mol Gen Genet 257:376–385
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kuhl JC, Hanneman RE, Havey MJ (2001) Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genomics 265:977–985
Lai Z, Livingstone K, Zou Y, Church SA, Knapp SJ, Andrews J, Rieseberg LH (2005a) Identification and mapping of SNPs from ESTs in sunflower. Theor Appl Genet 111:1532–1544
Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH (2005b) Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171:291–303
Leclercq P (1969) Cytoplasmic male sterility in sunflower. Ann Amelior Plant 19:99–106
Ling HQ, Qiu JW, Singh RP, Keller B (2004) Identification and genetic characterization of an Aegilops tauschii ortholog of the wheat leaf rust disease resistance gene Lr1. Theor Appl Genet 109:1133–1138
Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J (2003) High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82:378–389
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
Messeguer R, Ganal M, de Vicente MC, Young ND, Bolkan H, Tanksley SD (1991) High resolution RFLP map around the root knot nematode resistance gene (Mi) in tomato. Theor Appl Genet 82:529–536
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Micic Z, Hahn V, Bauer E, Schön CC, Melchinger AE (2005) QTL mapping of resistance to Sclerotinia midstalk rot in RIL of sunflower population NDBLOSsel × CM625. Theor Appl Genet 110:1490–1498
Miller JF, Gulya TJ (1991) Inheritance of resistance to race 4 of downy mildew derived from interspecific crosses in sunflower. Crop Sci 31:40–43
Miller JF, Seiler GJ (2003) Registration of five oilseed maintainer (HA 429-HA 433) sunflower germplasm lines. Crop Sci 43:2313–2314
Miller JF, Zimmerman DC, Vick BA, Roath WW (1987) Registration of sixteen high oleic sunflower germplasm lines and bulk population. Crop Sci 27:1323
Miller JF, Gulya TJ, Seiler GJ (2002) Registration of five fertility restorer sunflower germplasms. Crop Sci 42:989–991
Miller JF, Gulya TJ, Vick BA (2006) Registration of Imidazolinone herbicide-resistant maintainer (HA 442) and fertility restorer (RHA 443) oilseed sunflower germplasms. Crop Sci 46:483–484
Molinero-Ruiz ML, Melero-Vara JM (2003) First report of resistance to metalaxyl in downy mildew of sunflower caused by Plasmopara halstedii in Spain. Plant Dis 87:749
Molinero-Ruiz ML, Dominguez J, Melero-Vara JM (2002a) Races of isolates of Plasmopara halstedii from Spain and studies on their virulence. Plant Dis 86:736–740
Molinero-Ruiz ML, Melero-Vara JM, Dominguez J (2002b) Inheritance of resistance to race 330 of Plasmopara halstedii in three sunflower lines. Plant Breed 121:61–65
Molinero-Ruiz M, Melero-Vara J, Domínguez J (2003) Inheritance of resistance to two races of sunflower downy mildew (Plasmopara halstedii) in two Helianthus annuus L. lines. Euphytica 131:47–51
Molinero-Ruiz ML, Dominguez J, Gulya TJ, Melero-Vara JM (2005) Reaction of field populations of sunflower downy mildew (Plasmopara halstedii) to metalaxyl and mefenoxam. Helia 28:65–74
Mouzeyar S, Tourvieille De Labrouhe D, Vear F (1994) Effect of host-race combination on resistance of sunflower, Helianthus annuus L., to downy mildew Plasmopara halstedii. J Phytopathol 141:249–258
Mouzeyar S, Roeckel-Drevet P, Gentzbittel L, Philippon J, Tourvieille de Labroughe D, Vear F (1995) RFLP and RAPD mapping of the sunflower Pl1 locus for resistance to Plasmopara halstedii race 1. Theor Appl Genet 91:733–737
Mulpuri S, Liu Z, Feng J, Gulya T, Jan C-C (2009) Inheritance and molecular mapping of a downy mildew resistance gene, Pl 13 in cultivated sunflower (Helianthus annuus L.). Theor Appl Genet. doi:10.1007/s00122-00009-01089-z
Parlevliet J (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Radwan O, Bouzidi MF, Vear F, Philippon J, Tourvieille de Labrouhe D, Nicolas P, Mouzeyar S (2003) Identification of non-TIR-NBS-LRR markers linked to the Pl5/Pl8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Radwan O, Mouzeyar S, Venisse JS, Nicolas P, Bouzidi MF (2005) Resistance of sunflower to the biotrophic oomycete Plasmopara halstedii is associated with a delayed hypersensitive response within the hypocotyls. J Exp Bot 56:2683–2693
Radwan O, Gandhi S, Heesacker A, Whitaker B, Taylor C, Plocik A, Kesseli R, Kozik A, Michelmore RW, Knapp SJ (2008) Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol Genet Genomics 280:111–125
Rahim M, Jan CC, Gulya TJ (2002) Inheritance of resistance to sunflower downy mildew races 1, 2 and 3 in cultivated sunflower. Plant Breed 121:57–60
Roath WW, Miller JF, Gulya T (1987) Registration of three oilseed sunflower germplasm pools ND-BLPL2, ND-BLOS, and ND-RLOS. Crop Sci 27:373
Roeckel-Drevet P, Gagne G, Mouzeyar S, Gentzbittel L, Philippon J, Nicolas P, Tourvieille de Labrouhe D, Vear F (1996) Colocation of downy mildew (Plasmopara halstedii) resistance genes in sunflower (Helianthus annuus L.). Euphytica 91:225–228
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Sackston WE (1990) A proposed international system for designating races of Plasmopara halstedii. Plant Dis 74:721–723
Seah S, Yaghoobi J, Rossi M, Gleason CA, Williamson VM (2004) The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor Appl Genet 108:1635–1642
Seah S, Telleen A, Williamson V (2007) Introgressed and endogenous Mi-1 gene clusters in tomato differ by complex rearrangements in flanking sequences and show sequence exchange and diversifying selection among homologues. Theor Appl Genet 114:1289–1302
Seiler GJ (2007) Wild annual Helianthus anomalus and H. deserticola for improving oil content and quality in sunflower. Ind Crop Prod 25:95–100
Seiler GJ, Christie BR, Choo TM (1991) Registration of 13 downy mildew tolerant interspecific sunflower germplasm lines derived from wild annual species. Crop Sci 31:1714–1716
Singh KB, Ocampo B (1997) Exploitation of wild Cicer species for yield improvement in chickpea. Theor Appl Genet 95:418–423
Slabaugh MB, Huestis GM, Leonard J, Holloway JL, Rosato C, Hongtrakul V, Martini N, Toepfer R, Voetz M, Schell J, Knapp SJ (1997) Sequence-based genetic markers for genes and gene families: single-strand conformational polymorphisms for the fatty acid synthesis genes of Cuphea. Theor Appl Genet 94:400–408
Soderlund C, Longden I, Mott R (1997) FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13:523–535
Spring O, Zipper R, Heller-Dohmen M (2006) First report of metalaxyl resistant isolates of Plasmopara halstedii on cultivated sunflower in Germany. J Plant Dis Protect 113:224
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Tourvieille de Labrouhe D, Lafon S, Walser P, Raulic I (2000) Une nouvelle race de Plasmopara halstedii, agent du mildiou du tournesol. Oleagineux 7:404–405
Tourvieille de Labrouhe D, Serre F, Walser P, Roche S, Vear F (2008) Quantitative resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus). Euphytica 164:433–444
Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen
Vear F, Gentzbittel L, Philippon J, Mouzeyar S, Mestries E, Roeckel-Drevet P, Tourvieille de Labrouhe D, Nicolas P (1997) The genetics of resistance to five races of downy mildew (Plasmopara hastedii) in sunflower (Helianthus annuus L.). Theor Appl Genet 95:584–589
Vear F, Serre F, Roche S, Walser P, Tourvieille de Labrouhe D (2007) Recent research on downy midew resistance useful for breeding industrial-use sunflowers. Helia 30:45–54
Vear F, Serieys H, Petit A, Serre F, Boudon J-P, Roche S, Walser P, Tourvieille de Labrouhe D (2008a) Origins of major genes for downy mildew resistance in sunflower. In: Velasco I (ed) Proceedings of 17th international sunflower conference, Córdoba, Spain, pp 125–130
Vear F, Serre F, Jouan-Dufournel I, Bert PF, Roche S, Walser P, Tourvieille de Labrouhe D, Vincourt P (2008b) Inheritance of quantitative resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Euphytica 164:561–570
Vranceanu V, Stoenescu F (1970) Immunity to sunflower downy mildew due to a single dominant gene. Probl Agric 22:34–40
Vranceanu VL, Pirvu N, Stoenescu FM (1981) New sunflower downy mildew resistance genes and their management. Helia 4:23–27
Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP (1999) The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153:1929–1948
Yu J-K, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M, Soper J, Han F, Chu W-C, Webb DM, Thompson L, Edwards KJ, Berry S, Leon AJ, Grondona M, Olungu C, Maes N, Knapp SJ (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Zheng J, Svensson J, Madishetty K, Close T, Jiang T, Lonardi S (2006) OligoSpawn: a software tool for the design of overgo probes from large unigene datasets. BMC Bioinform 7:7
Zimmer DE, Kinman ML (1972) Downy mildew resistance in cultivated sunflower and its inheritance. Crop Sci 12:749–751
Acknowledgments
Sunflower seeds of cmsHA342 x ARG1575-2 were kindly provided by Prof. Dr. W. Friedt, Justus-Liebig University, Gießen, Germany. We thank Dr. G. Seiler, U.S. Department of Agriculture, Agricultural Research Service, Fargo, for seeds of RHA419, RHA420 and RHA443, and Dr. André Bervillé, INRA, Montpellier, for seeds of ARG79MTP and FS20-6-2. We also thank Prof. Dr. O. Spring, University of Hohenheim, Stuttgart, Germany, for providing us with the P. halstedii races 100, 330, and 710. Many thanks belong to Stefan Schwertfirm, Barbara Renz, Anna Elhabachi, and Sabine Schillinger for technical assistance. This study is supported by a grant (no. BA 2073/2-4) from the Deutsche Forschungsgemeinschaft and a scholarship for Silke Wieckhorst from the Universität Bayern e.V.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wieckhorst, S., Bachlava, E., Dußle, C.M. et al. Fine mapping of the sunflower resistance locus Pl ARG introduced from the wild species Helianthus argophyllus . Theor Appl Genet 121, 1633–1644 (2010). https://doi.org/10.1007/s00122-010-1416-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1416-4