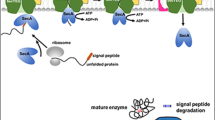
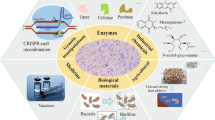
Abstract
Bacillus licheniformis is a characteristic Gram-positive bacterium originally found in soil. This microorganism has long been utilised as a workhorse for production of industrial enzymes or high value-added chemicals. With ever-increasing understanding on this strain and the maturation of the genetic technique, important advances have recently been made in developing B. licheniformis as an excellent chassis cell for synthetic biology. Here, we provide an overview of updated understanding on genome information, anaerobic metabolism, industrial applications of this strain. The state-of-art B. licheniformis genetics, especially its synthetic biology advances in biosensor, expression system and artificial metabolic pathways are illustrated. Finally, perspectives are offered for the limitations and challenges to be addressed to improve B. licheniformis as microbial cell factories.




Similar content being viewed by others
Data availability
Not applicable.
References
Xu Y, Li Y, Wu Z, Lu Y, Tao G, Zhang L, Ding Z, Shi G. Combining precursor-directed engineering with modular designing: an effective strategy for de novo biosynthesis of l-DOPA in Bacillus licheniformis. ACS Synth Biol. 2022;11(2):700–12. https://doi.org/10.1021/acssynbio.1c00411.
Wu Z, Li Y, Xu Y, Zhang Y, Tao G, Zhang L, Shi G. Transcriptome analysis of Bacillus licheniformis for improving bacitracin production. ACS Synth Biol. 2022. https://doi.org/10.1021/acssynbio.1c00593.
Voigt B, Antelmann H, Albrecht D, Ehrenreich A, Maurer KH, Evers S, Gottschalk G, van Dijl JM, Schweder T, Hecker M. Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis. J Mol Microbiol Biotechnol. 2009;16(1–2):53–68. https://doi.org/10.1159/000142894.
Lv X, Hueso-Gil A, Bi X, Wu Y, Liu Y, Liu L, Ledesma-Amaro R. New synthetic biology tools for metabolic control. Curr Opin Biotechnol. 2022;76:102724. https://doi.org/10.1016/j.copbio.2022.102724.
Muras A, Romero M, Mayer C, Otero A. Biotechnological applications of Bacillus licheniformis. Crit Rev Biotechnol. 2021;41(4):609–27. https://doi.org/10.1080/07388551.2021.1873239.
Skerman VMVBD, Sneath PHA. Approved lists of bacterial names. Washington: The Ad Hoc Committee of the Judicial Commission of the ICSB; 1980.
Rey MW, Ramaiya P, Nelson BA, Brody-Karpin SD, Zaretsky EJ, Tang M, de Leon AL, Xiang H, Gusti V, Clausen IG, Olsen PB, Rasmussen MD, Andersen JT, Jorgensen PL, Larsen TS, Sorokin A, Bolotin A, Lapidus A, Galleron N, Ehrlich SD, Berka RM. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 2004;5(10):r77. https://doi.org/10.1186/gb-2004-5-10-r77.
Dindhoria K, Sanjeet K, Baliyan N, Raphel S, Halami PM, Kumar R. Bacillus licheniformis MCC 2514 genome sequencing and functional annotation for providing genetic evidence for probiotic gut adhesion properties and its applicability as a bio-preservative agent. Gene. 2022;840:146744. https://doi.org/10.1016/j.gene.2022.146744.
Rachinger M, Volland S, Meinhardt F, Daniel R, Liesegang H. First insights into the completely annotated genome sequence of Bacillus licheniformis strain 9945A. Genome Announc. 2013. https://doi.org/10.1128/genomeA.00525-13.
Yang F, Liu Y, Chen L, Li J, Wang L, Du G. Genome sequencing and flavor compound biosynthesis pathway analyses of Bacillus licheniformis isolated from Chinese Maotai-flavor liquor-brewing microbiome. Food Biotechnol. 2020;34(3):193–211. https://doi.org/10.1080/08905436.2020.1789474.
Arora PK, Mishra R, Omar RA, Saroj RS, Srivastava A, Garg SK, Singh VP. Draft genome sequence data of a chromium reducing bacterium, Bacillus licheniformis strain KNP. Data Brief. 2021;34:106640. https://doi.org/10.1016/j.dib.2020.106640.
Borriss R. Bacillus, beneficial microbes in agro-ecology. Amsterdam: Elsevier; 2020. p. 107–32.
Esmaeilishirazifard E, De Vizio D, Moschos SA, Keshavarz T. Genomic and molecular characterization of a novel quorum sensing molecule in Bacillus licheniformis. AMB Express. 2017;7(1):78. https://doi.org/10.1186/s13568-017-0381-6.
Lee C, Kim JY, Song HS, Kim YB, Choi YE, Yoon C, Nam YD, Roh SW. Genomic analysis of Bacillus licheniformis CBA7126 isolated from a human fecal sample. Front Pharmacol. 2017;8:724. https://doi.org/10.3389/fphar.2017.00724.
Sharma A, Satyanarayana T. Comparative genomics of Bacillus species and its relevance in industrial microbiology. Genomics Insights. 2013;6:25–36. https://doi.org/10.4137/GEI.S12732.
Dunlap CA, Kwon SW, Rooney AP, Kim SJ. Bacillus paralicheniformis sp. Nov., isolated from fermented soybean paste. Int J Syst Evol Microbiol. 2015;65(10):3487–92. https://doi.org/10.1099/ijsem.0.000441.
Olajide AM, Chen S, LaPointe G. Markers to rapidly distinguish Bacillus paralicheniformis from the very close relative, Bacillus licheniformis. Front Microbiol. 2021. https://doi.org/10.3389/fmicb.2020.596828.
Clements LD, Miller BS, Streips UN. Comparative growth analysis of the facultative anaerobes Bacillus subtilis, Bacillus licheniformis, and Escherichia coli. Syst Appl Microbiol. 2002;25(2):284–6. https://doi.org/10.1078/0723-2020-00108.
Maghnouj A, Abu-Bakr AA, Baumberg S, Stalon V, Vander Wauven C. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol Lett. 2000;191(2):227–34. https://doi.org/10.1111/j.1574-6968.2000.tb09344.x.
Wohlkonig A, Stalon V, Vander Wauven C. Purification of ArcR, an oxidation-sensitive regulatory protein from Bacillus licheniformis. Protein Expr Purif. 2004;37(1):32–8. https://doi.org/10.1016/j.pep.2004.05.006.
Klinger A, Schirawski J, Glaser P, Unden G. The fnr gene of Bacillus licheniformis and the cysteine ligands of the C-terminal FeS cluster. J Bacteriol. 1998;180(13):3483–5. https://doi.org/10.1128/JB.180.13.3483-3485.1998.
Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Baumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol. 2004;7(4):204–11. https://doi.org/10.1159/000079829.
Shariati P, Mitchell WJ, Boyd A, Priest FG. Anaerobic metabolism in Bacillus-licheniformis Ncib-6346. Microbiology-UK. 1995;141:1117–24. https://doi.org/10.1099/13500872-141-5-1117.
Gudina EJ, Teixeira JA. Bacillus licheniformis: the unexplored alternative for the anaerobic production of lipopeptide biosurfactants? Biotechnol Adv. 2022;60:108013. https://doi.org/10.1016/j.biotechadv.2022.108013.
Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56(4):845–57. https://doi.org/10.1111/j.1365-2958.2005.04587.x.
Willenbacher MZJ, Mohr T, Schmidt F, Syldatk C, Hausmann R. Evaluation of different Bacillus strains in respect of their ability to produce surfactin in a model fermentation process with integrated foam fractionation. Appl Microbiol Biotechnol. 2014;98:9623–32. https://doi.org/10.1007/s00253-014-6010-2.
Harwood CR, Mouillon JM, Pohl S, Arnau J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol Rev. 2018;42(6):721–38. https://doi.org/10.1093/femsre/fuy028.
Zhang R, Wu Q, Xu Y. Lichenysin, a cyclooctapeptide occurring in Chinese liquor Jian Nan Chun reduced the headspace concentration of phenolic off-flavors via hydrogen-bond interactions. J Agric Food Chem. 2014;62(33):8302–7. https://doi.org/10.1021/jf502053g.
Coronel-Leon J, Marques AM, Bastida J, Manresa A. Optimizing the production of the biosurfactant lichenysin and its application in biofilm control. J Appl Microbiol. 2016;120(1):99–111. https://doi.org/10.1111/jam.12992.
Batrakov SG, Rodionova TA, Esipov SE, Polyakov NB, Sheichenko VI, Shekhovtsova NV, Lukin SM, Panikov NS, Nikolaev YA. A novel lipopeptide, an inhibitor of bacterial adhesion, from the thermophilic and halotolerant subsurface Bacillus licheniformis strain 603. BBA-Mol Cell Biol Lipids. 2003;1634(3):107–15. https://doi.org/10.1016/j.bbalip.2003.09.004.
Joshi SYS, Desai AJ. Application of response-surface methodology to evaluate the optimum medium components for the enhanced production of lichenysin by Bacillus licheniformis R2. Biochem Eng J. 2008;41(2):122–7. https://doi.org/10.1016/j.bej.2008.04.005.
Ali N, Wang F, Xu B, Safdar B, Ullah A, Naveed M, Wang C, Rashid MT. Production and application of biosurfactant produced by Bacillus Licheniformis Ali5 in enhanced oil recovery and motor oil removal from contaminated sand. Molecules. 2019;24(24):4448. https://doi.org/10.3390/molecules24244448.
Qiu Y, Xiao F, Wei X, Wen Z, Chen S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol. 2014;98(21):8895–903. https://doi.org/10.1007/s00253-014-5978-y.
Lin KGLSC, Lo CC, Lin YM. Enhanced biosurfactant production by a Bacillus licheniformis mutant. Enzym Microb Technol. 1998;23:267–73. https://doi.org/10.1016/S0141-0229(98)00049-0.
Zhu C, Xiao F, Qiu Y, Wang Q, He Z, Chen S. Lichenysin production is improved in codY null Bacillus licheniformis by addition of precursor amino acids. Appl Microbiol Biotechnol. 2017;101(16):6375–83. https://doi.org/10.1007/s00253-017-8352-z.
Sun F, Yu DZ, Zhou HY, Lin HK, Yan Z, Wu AB. CotA laccase from Bacillus licheniformis ZOM-1 effectively degrades zearalenone, aflatoxin B1 and alternariol. Food Control. 2022;145:109472. https://doi.org/10.1016/j.foodcont.2022.109472.
Chen YH, Zhang RZ, Zhang WC, Xu Y. Alanine aminopeptidase from Bacillus licheniformis E7 expressed in Bacillus subtilis efficiently hydrolyzes soy protein to small peptides and free amino acids. LWT Food Sci Technol. 2022;165:113642. https://doi.org/10.1016/j.lwt.2022.113642.
Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50(1):1–17. https://doi.org/10.1139/W03-076.
Niu DD, Zuo ZR, Shi GY, Wang ZX. High yield recombinant thermostable alpha-amylase production using an improved Bacillus licheniformis system. Microb Cell Factories. 2009. https://doi.org/10.1186/1475-2859-8-58.
Craynest M, Jorgensen S, Sarvas M, Kontinen VP. Enhanced secretion of heterologous cyclodextrin glycosyltransferase by a mutant of Bacillus licheniformis defective in the D-alanylation of teichoic acids. Lett Appl Microbiol. 2003;37(1):75–80. https://doi.org/10.1046/j.1472-765X.2003.01357.x.
Ruan YQ, Xu Y, Zhang WC, Zhang RZ. A new maltogenic amylase from Bacillus licheniformis R-53 significantly improves bread quality and extends shelf life. Food Chem. 2021;344:128599. https://doi.org/10.1016/j.foodchem.2020.128599.
Silva-Salinas A, Rodriguez-Delgado M, Gomez-Trevino J, Lopez-Chuken U, Olvera-Carranza C, Blanco-Gamez EA. Novel thermotolerant amylase from Bacillus licheniformis strain LB04: purification, characterization and agar-agarose. Microorganisms. 2021;9(9):1857. https://doi.org/10.3390/microorganisms9091857.
Fan Y, Wang J, Gao C, Zhang Y, Du W. A novel exopolysaccharide-producing and long-chain n-alkane degrading bacterium Bacillus licheniformis strain DM-1 with potential application for in-situ enhanced oil recovery. Sci Rep. 2020;10(1):8519. https://doi.org/10.1038/s41598-020-65432-z.
Gao Z. Purification and characterization of a novel lichenase from Bacillus licheniformis GZ-2. Biotechnol Appl Biochem. 2016;63(2):249–56. https://doi.org/10.1002/bab.1206.
Kim HW, Lee DC, Rhee HI. Production of alpha-glucosidase inhibitor in the intestines by Bacillus licheniformis. Enzyme Microb Technol. 2022;158:110032. https://doi.org/10.1016/j.enzmictec.2022.110032.
Meng W, Ma C, Xu P, Gao C. Biotechnological production of chiral acetoin. Trends Biotechnol. 2022;40(8):958–73. https://doi.org/10.1016/j.tibtech.2022.01.008.
Wu Y, Liu Y, Lv X, Li J, Du G, Liu L. CAMERS-B: CRISPR/Cpf1 assisted multiple-genes editing and regulation system for Bacillus subtilis. Biotechnol Bioeng. 2020;117(6):1817–25. https://doi.org/10.1002/bit.27322.
Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. Metabolic engineering strategies for acetoin and 2,3-butanediol production: advances and prospects. Crit Rev Biotechnol. 2017;37(8):990–1005. https://doi.org/10.1080/07388551.2017.1299680.
Liu YF, Zhang SL, Yong YC, Ji ZX, Ma X, Xu ZH, Chen SW. Efficient production of acetoin by the newly isolated Bacillus licheniformis strain MEL09. Process Biochem. 2011;46(1):390–4. https://doi.org/10.1016/j.procbio.2010.07.024.
Li L, Wei XT, Yu WP, Wen ZY, Chen SW. Enhancement of acetoin production from Bacillus licheniformis by 2,3-butanediol conversion strategy: metabolic engineering and fermentation control. Process Biochem. 2017;57:35–42. https://doi.org/10.1016/j.procbio.2017.03.027.
Zhang X, Zhang RZ, Bao T, Rao ZM, Yang TW, Xu MJ, Xu ZH, Li HZ, Yang ST. The rebalanced pathway significantly enhances acetoin production by disruption of acetoin reductase gene and moderate-expression of a new water-forming NADH oxidase in Bacillus subtilis. Metab Eng. 2014;23:34–41. https://doi.org/10.1016/j.ymben.2014.02.002.
Luo QL, Wu J, Wu MC. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem. 2014;49(8):1223–30. https://doi.org/10.1016/j.procbio.2014.05.005.
Yuan HL, Xu Y, Chen YZ, Zhan YY, Wei XT, Li L, Wang D, He PH, Li SQ, Chen SW. Metabolomics analysis reveals global acetoin stress response of Bacillus licheniformis. Metabolomics. 2019. https://doi.org/10.1007/s11306-019-1492-7.
Kim JW, Seo SO, Zhang GC, Jin YS, Seo JH. Expression of Lactococcus lactis NADH oxidase increases 2,3-butanediol production in Pdc-deficient Saccharomyces cerevisiae. Bioresour Technol. 2015;191:512–9. https://doi.org/10.1016/j.biortech.2015.02.077.
Kim S, Hahn JS. Synthetic scaffold based on a cohesin-dockerin interaction for improved production of 2,3-butanediol in Saccharomyces cerevisiae. J Biotechnol. 2014;192:192–6. https://doi.org/10.1016/j.jbiotec.2014.10.015.
Thanh TN, Jurgen B, Bauch M, Liebeke M, Lalk M, Ehrenreich A, Evers S, Maurer KH, Antelmann H, Ernst F, Homuth G, Hecker M, Schweder T. Regulation of acetoin and 2,3-butanediol utilization in Bacillus licheniformis. Appl Microbiol Biotechnol. 2010;87(6):2227–35. https://doi.org/10.1007/s00253-010-2681-5.
Kabisch J, Pratzka I, Meyer H, Albrecht D, Lalk M, Ehrenreich A, Schweder T. Metabolic engineering of Bacillus subtilis for growth on overflow metabolites. Microb Cell Fact. 2013;12:72. https://doi.org/10.1186/1475-2859-12-72.
Jiang M, Li Q, Hu S, He P, Chen Y, Cai D, Wu Y, Chen S. Enhanced aerobic denitrification performance with Bacillus licheniformis via secreting lipopeptide biosurfactant lichenysin. Chem Eng J. 2022;434:134686. https://doi.org/10.1016/j.cej.2022.134686.
Yang Q, Yang T, Shi Y, Xin Y, Zhang L, Gu ZH, Li YR, Ding ZY, Shi GY. The nitrogen removal characterization of a cold-adapted bacterium: Bacillus simplex H-b. Bioresour Technol. 2021;323:124554. https://doi.org/10.1016/j.biortech.2020.124554.
Takenaka S, Zhou Q, Kuntiya A, Seesuriyachan P, Murakami S, Aoki K. Isolation and characterization of thermotolerant bacterium utilizing ammonium and nitrate ions under aerobic conditions. Biotechnol Lett. 2007;29(3):385–90. https://doi.org/10.1007/s10529-006-9255-8.
Liang Q, Zhang X, Lee KH, Wang Y, Yu K, Shen W, Fu L, Shu M, Li W. Nitrogen removal and water microbiota in grass carp culture following supplementation with Bacillus licheniformis BSK-4. World J Microbiol Biotechnol. 2015;31(11):1711–8. https://doi.org/10.1007/s11274-015-1921-3.
Vasconcellos SP, Cereda MP, Cagnon JR, Foglio MA, Rodrigues RA, Manfio GP, Oliveira VM. In vitro degradation of linamarin by microorganisms isolated from cassava wastewater treatment lagoons. Braz J Microbiol. 2009;40(4):879–83. https://doi.org/10.1590/S1517-838220090004000019.
Ye S, Zeng G, Wu H, Zhang C, Dai J, Liang J, Yu J, Ren X, Yi H, Cheng M, Zhang C. Biological technologies for the remediation of co-contaminated soil. Crit Rev Biotechnol. 2017;37(8):1062–76. https://doi.org/10.1080/07388551.2017.1304357.
Xu NN, Bao MT, Sun PY, Li YM. Study on bioadsorption and biodegradation of petroleum hydrocarbons by a microbial consortium. Bioresour Technol. 2013;149:22–30. https://doi.org/10.1016/j.biortech.2013.09.024.
Thion C, Cébron A, Beguiristain T, Leyval C. PAH biotransformation and sorption by Fusarium solani and Arthrobacter oxydans isolated from a polluted soil in axenic cultures and mixed co-cultures. Int Biodeterior Biodegrad. 2012;68:28–35. https://doi.org/10.1016/j.ibiod.2011.10.012.
Deive FJ, Carvalho E, Pastrana L, Rua ML, Longo MA, Sanroman MA. Strategies for improving extracellular lipolytic enzyme production by Thermus thermophilus HB27. Bioresour Technol. 2009;100(14):3630–7. https://doi.org/10.1016/j.biortech.2009.02.053.
Jamil M, Zeb S, Anees M, Roohi A, Ahmed I, ur Rehman S, Rha ES. Role of Bacillus licheniformis in phytoremediation of nickel contaminated soil cultivated with rice. Int J Phytoremediat. 2014;16(6):554–71. https://doi.org/10.1080/15226514.2013.798621.
Upadhyay KH, Vaishnav AM, Tipre DR, Patel BC, Dave SR. Kinetics and mechanisms of mercury biosorption by an exopolysaccharide producing marine isolate Bacillus licheniformis. 3 Biotech. 2017;7(5):313. https://doi.org/10.1007/s13205-017-0958-4.
Chris Felshia S, Aswin Karthick N, Thilagam R, Chandralekha A, Raghavarao K, Gnanamani A. Efficacy of free and encapsulated Bacillus lichenformis strain SL10 on degradation of phenol: a comparative study of degradation kinetics. J Environ Manag. 2017;197:373–83. https://doi.org/10.1016/j.jenvman.2017.04.005.
Shi S, Xie Y, Wang G, Luo Y. Metabolite-based biosensors for natural product discovery and overproduction. Curr Opin Biotechnol. 2022;75:102699. https://doi.org/10.1016/j.copbio.2022.102699.
Teng Y, Zhang J, Jiang T, Zou Y, Gong X, Yan Y. Biosensor-enabled pathway optimization in metabolic engineering. Curr Opin Biotechnol. 2022;75:102696. https://doi.org/10.1016/j.copbio.2022.102696.
Qin L, Liu X, Xu K, Li C. Mining and design of biosensors for engineering microbial cell factory. Curr Opin Biotechnol. 2022;75:102694. https://doi.org/10.1016/j.copbio.2022.102694.
Gao C, Hou J, Xu P, Guo L, Chen X, Hu G, Ye C, Edwards H, Chen J, Chen W, Liu L. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat Commun. 2019;10(1):3751. https://doi.org/10.1038/s41467-019-11793-7.
Zou X, Cheng C, Feng J, Song X, Lin M, Yang ST. Biosynthesis of polymalic acid in fermentation: advances and prospects for industrial application. Crit Rev Biotechnol. 2019;39(3):408–21. https://doi.org/10.1080/07388551.2019.1571008.
Zhang Y, Li Y, Xiao F, Wang H, Zhang L, Ding Z, Xu S, Gu Z, Shi G. Engineering of a biosensor in response to Malate in Bacillus licheniformis. ACS Synth Biol. 2021;10(7):1775–84. https://doi.org/10.1021/acssynbio.1c00170.
Wu H, Liu S, Jiang J, Shen G, Yu R. A novel electrochemical biosensor for highly selective detection of protease biomarker from Bacillus licheniformis with D-amino acid containing peptide. Analyst. 2012;137(20):4829–33. https://doi.org/10.1039/c2an36066g.
Au HW, Tsang MW, So PK, Wong KY, Leung YC. Thermostable beta-lactamase mutant with its active site conjugated with fluorescein for efficient beta-lactam antibiotic detection. ACS Omega. 2019;4(24):20493–502. https://doi.org/10.1021/acsomega.9b02211.
Li Y, Jin K, Zhang L, Ding Z, Gu Z, Shi G. Development of an inducible secretory expression system in Bacillus licheniformis based on an engineered xylose operon. J Agric Food Chem. 2018;66(36):9456–64. https://doi.org/10.1021/acs.jafc.8b02857.
Yang MM, Zhang WW, Ji SY, Cao PH, Chen YL, Zhao X. Generation of an artificial double promoter for protein expression in Bacillus subtilis through a promoter trap system. PLoS ONE. 2013. https://doi.org/10.1371/journal.pone.0056321.
Shen P, Niu D, Permaul K, Tian K, Singh S, Wang Z. Exploitation of ammonia-inducible promoters for enzyme overexpression in Bacillus licheniformis. J Ind Microbiol Biotechnol. 2021. https://doi.org/10.1093/jimb/kuab037.
Trung NT, Hung NM, Thuan NH, Canh NX, Schweder T, Jurgen B. An auto-inducible phosphate-controlled expression system of Bacillus licheniformis. BMC Biotechnol. 2019;19(1):3. https://doi.org/10.1186/s12896-018-0490-6.
Zhan YY, Xu Y, Lu XC, Zhou F, Zheng PL, Wang D, Cai DB, Yang SH, Chen SW. Metabolic engineering of Bacillus licheniformis for sustainable production of isobutanol. Acs Sustain Chem Eng. 2021;9(51):17254–65. https://doi.org/10.1021/acssuschemeng.1c05511.
Li YR, Ma XF, Zhang L, Ding ZY, Xu S, Gu ZH, Shi GY. Engineering of Bacillus promoters based on interacting motifs between UP elements and RNA polymerase (RNAP) alpha-subunit. Int J Mol Sci. 2022;23(21):13480. https://doi.org/10.3390/ijms232113480.
Xiao F, Li Y, Zhang Y, Wang H, Zhang L, Ding Z, Gu Z, Xu S, Shi G. A new CcpA binding site plays a bidirectional role in carbon catabolism in Bacillus licheniformis. Iscience. 2021;24(5):102400. https://doi.org/10.1016/j.isci.2021.102400.
Hertel R, Volland S, Liesegang H. Conjugative reporter system for the use in Bacillus licheniformis and closely related Bacilli. Lett Appl Microbiol. 2015;60(2):162–7. https://doi.org/10.1111/lam.12352.
Wang S, Wang H, Zhang D, Li X, Zhu J, Zhan Y, Cai D, Wang Q, Ma X, Wang D, Chen S. Multistep metabolic engineering of Bacillus licheniformis to improve pulcherriminic acid production. Appl Environ Microbiol. 2020;86:9. https://doi.org/10.1128/aem.03041-19.
Zhan Y, Xu Y, Zheng P, He M, Sun S, Wang D, Cai D, Ma X, Chen S. Establishment and application of multiplexed CRISPR interference system in Bacillus licheniformis. Appl Microbiol Biotechnol. 2020;104(1):391–403. https://doi.org/10.1007/s00253-019-10230-5.
Qiu Y, Zhang J, Li L, Wen Z, Nomura CT, Wu S, Chen S. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol Biofuels. 2016;9:117. https://doi.org/10.1186/s13068-016-0522-1.
Mo F, Cai D, He P, Yang F, Chen Y, Ma X, Chen S. Enhanced production of heterologous proteins via engineering the cell surface of Bacillus licheniformis. J Ind Microbiol Biotechnol. 2019;46(12):1745–55. https://doi.org/10.1007/s10295-019-02229-8.
Wang J, Zhu L, Li Y, Xu S, Jiang W, Liang C, Fang Y, Chu A, Zhang L, Ding Z, Shi G. Enhancing geranylgeraniol production by metabolic engineering and utilization of isoprenol as a substrate in Saccharomyces cerevisiae. J Agric Food Chem. 2021;69(15):4480–9. https://doi.org/10.1021/acs.jafc.1c00508.
Li Y, Wang H, Zhang L, Ding Z, Xu S, Gu Z, Shi G. Efficient genome editing in Bacillus licheniformis mediated by a conditional CRISPR/Cas9 system. Microorganisms. 2020;8(5):754. https://doi.org/10.3390/microorganisms8050754.
Li KF, Cai DB, Wang ZQ, He ZL, Chen SW. Development of an efficient genome editing tool in Bacillus licheniformis using CRISPR-Cas9 nickase. Appl Environ Microbiol. 2018. https://doi.org/10.1128/AEM.02608-17.
Acknowledgements
This work was supported by National Key Research & Development Program of China (2018YFA0900504, 2020YFA0907700, and 2018YFA0900300), the National Natural Foundation of China (31401674), the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-22), and the Top-notch Academic Programs Project of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing financial interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, H., Zhang, Y., Shi, G. et al. Recent biotechnological advances and future prospective of Bacillus licheniformis as microbial cell factories. Syst Microbiol and Biomanuf 3, 521–532 (2023). https://doi.org/10.1007/s43393-023-00162-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-023-00162-7