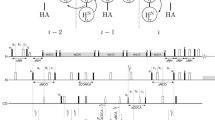
Abstract
Sequence specific resonance assignment is the prerequisite for the NMR-based analysis of the conformational ensembles and their underlying dynamics of intrinsically disordered proteins. However, rapid solvent exchange in intrinsically disordered proteins often complicates assignment strategies based on HN-detection. Here we present a six-dimensional alpha proton detection-based automated projection spectroscopy (APSY) experiment for backbone assignment of intrinsically disordered proteins. The 6D HCACONCAH APSY correlates the six different chemical shifts, Hα(i − 1), Cα(i − 1), C′(i − 1), N(i), Cα(i) and Hα(i). Application to two intrinsically disordered proteins, 140-residue α-synuclein and a 352-residue isoform of Tau, demonstrates that the chemical shift information provided by the 6D HCACONCAH APSY allows efficient backbone resonance assignment of intrinsically disordered proteins.






Similar content being viewed by others
References
Atreya HS, Szyperski T (2004) G-matrix Fourier transform NMR spectroscopy for complete protein resonance assignment. Proc Natl Acad Sci USA 101:9642–9647. doi:10.1073/pnas.0403529101
Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP (1987) Exponential sampling, an alternative method for sampling in two-dimensional NMR experiments. J Magn Reson 73:69–77. doi:10.1016/0022-2364(87)90225-3
Bermel W, Bertini I, Felli IC, Kummerle R, Pierattelli R (2006a) Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J Magn Reson 178:56–64. doi:10.1016/j.jmr.2005.08.011
Bermel W, Bertini I, Felli IC, Lee YM, Luchinat C, Pierattelli R (2006b) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc 128:3918–3919. doi:10.1021/Ja0582206
Bermel W, Bertini I, Chill J, Felli IC, Haba N, Kumar MVV, Pierattelli R (2012a) Exclusively heteronuclear C-13-Detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDPs). Chembiochem 13:2425–2432. doi:10.1002/cbic.201200447
Bermel W et al (2012b) Speeding up sequence specific assignment of IDPs. J Biomol NMR 53:293–301. doi:10.1007/s10858-012-9639-0
Bermel W, Felli IC, Gonnelli L, Kozminski W, Piai A, Pierattelli R, Zawadzka-Kazimierczuk A (2013) High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR 57:353–361. doi:10.1007/s10858-013-9793-z
Bertini I, Duma L, Felli IC, Fey M, Luchinat C, Pierattelli R, Vasos PR (2004) A heteronuclear direct-detection NMR spectroscopy experiment for protein-backbone assignment. Angew Chem Int Edit 43:2257–2259. doi:10.1002/anie.200453661
Bottomley MJ, Macias MJ, Liu Z, Sattler M (1999) A novel NMR experiment for the sequential assignment of proline residues and proline stretches in 13C/15 N-labeled proteins. J Biomol NMR 13:381–385
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Chylla RA, Markley JL (1995) Theory and application of the maximum likelihood principle to NMR parameter estimation of multidimensional NMR data. J Biomol NMR 5:245–258
Coggins BE, Zhou P (2006) Polar Fourier transforms of radially sampled NMR data. J Magn Reson 182:84–95. doi:10.1016/j.jmr.2006.06.016
Coggins BE, Venters RA, Zhou P (2004) Generalized reconstruction of n-D NMR spectra from multiple projections: application to the 5-D HACACONH spectrum of protein G B1 domain. J Am Chem Soc 126:1000–1001. doi:10.1021/ja039430q
Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT (2008) Hydrogen exchange of monomeric alpha-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci 17:1434–1445. doi:10.1110/ps.033803.107
Csizmok V, Felli IC, Tompa P, Banci L, Bertini I (2008) Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Am Chem Soc 130:16873–16879. doi:10.1021/ja805510b
Felli IC, Brutscher B (2009) Recent advances in solution NMR: fast methods and heteronuclear direct detection. Chemphyschem 10:1356–1368. doi:10.1002/cphc.200900133
Fiorito F, Hiller S, Wider G, Wuthrich K (2006) Automated resonance assignment of proteins: 6D APSY-NMR. J Biomol NMR 35:27–37. doi:10.1007/s10858-006-0030-x
Güntert P, Dötsch V, Wider G, Wüthrich K (1992) Processing of multidimensional NMR data with the new software PROSA. J Biomol NMR 2:619–629. doi:10.1007/Bf02192850
Hiller S, Fiorito F, Wuthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102:10876–10881. doi:10.1073/pnas.0504818102
Hiller S, Wider G, Wuthrich K (2008) APSY-NMR with proteins: practical aspects and backbone assignment. J Biomol NMR 42:179–195. doi:10.1007/s10858-008-9266-y
Holland DJ, Bostock MJ, Gladden LF, Nietlispach D (2011) Fast multidimensional NMR spectroscopy using compressed sensing. Angew Chem Int Edit 50:6548–6551. doi:10.1002/Anie.201100440
Hu K, Vogeli B, Clore GM (2007) Spin-state selective carbon-detected HNCO with TROSY optimization in all dimensions and double echo-antiecho sensitivity enhancement in both indirect dimensions. J Am Chem Soc 129:5484–5491. doi:10.1021/ja067981l
Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK (2002) Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323:573–584
Jensen MR, Ruigrok RW, Blackledge M (2013) Describing intrinsically disordered proteins at atomic resolution by NMR. Curr Opin Struct Biol 23:426–435. doi:10.1016/j.sbi.2013.02.007
Jung YS, Zweckstetter M (2004) Mars—robust automatic backbone assignment of proteins. J Biomol NMR 30:11–23. doi:10.1023/B:JNMR.0000042954.99056.ad
Kanelis V, Donaldson L, Muhandiram DR, Rotin D, Forman-Kay JD, Kay LE (2000) Sequential assignment of proline-rich regions in proteins: application to modular binding domain complexes. J Biomol NMR 16:253–259. doi:10.1023/A:1008355012528
Kay LE, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114:10663–10665. doi:10.1021/Ja00052a088
Kazimierczuk K, Orekhov VY (2011) Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed Engl 50:5556–5559. doi:10.1002/anie.201100370
Kazimierczuk K, Kozminski W, Zhukov I (2006) Two-dimensional Fourier transform of arbitrarily sampled NMR data sets. J Magn Reson 179:323–328. doi:10.1016/j.jmr.2006.02.001
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W (2013) High-dimensional NMR spectra for structural studies of biomolecules. Chemphyschem 14:3015–3025. doi:10.1002/cphc.201300277
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125:1385–1393. doi:10.1021/Ja028197d
Korzhneva DM, Ibraghimov IV, Billeter M, Orekhov VY (2001) MUNIN: application of three-way decomposition to the analysis of heteronuclear NMR relaxation data. J Biomol NMR 21:263–268
Kupce E, Freeman R (2003a) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125:13958–13959. doi:10.1021/Ja038297z
Kupce E, Freeman R (2003b) Reconstruction of the three-dimensional NMR spectrum of a protein from a set of plane projections. J Biomol NMR 27:383–387
Kupce E, Freeman R (2004) Projection-reconstruction technique for speeding up multidimensional NMR spectroscopy. J Am Chem Soc 126:6429–6440. doi:10.1021/ja049432q
Mantylahti S, Aitio O, Hellman M, Permi P (2010) HA-detected experiments for the backbone assignment of intrinsically disordered proteins. J Biomol NMR 47:171–181. doi:10.1007/s10858-010-9421-0
Mantylahti S, Hellman M, Permi P (2011) Extension of the HA-detection based approach: (HCA)CON(CA)H and (HCA)NCO(CA)H experiments for the main-chain assignment of intrinsically disordered proteins. J Biomol NMR 49:99–109. doi:10.1007/s10858-011-9470-z
Marsh JA, Forman-Kay JD (2010) Sequence determinants of compaction in intrinsically disordered proteins. Biophys J 98:2383–2390. doi:10.1016/j.bpj.2010.02.006
Mccoy MA, Mueller L (1992) Nonresonant effects of frequency-selective pulses. J Magn Reson 99:18–36. doi:10.1016/0022-2364(92)90152-W
Mittag T, Forman-Kay JD (2007) Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol 17:3–14. doi:10.1016/j.sbi.2007.01.009
Motackova V et al (2010) Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J Biomol NMR 48:169–177. doi:10.1007/s10858-010-9447-3
Narayanan RL, Durr UH, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132:11906–11907. doi:10.1021/ja105657f
Novacek J et al (2011) 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR 50:1–11. doi:10.1007/s10858-011-9496-2
Novacek J, Janda L, Dopitova R, Zidek L, Sklenar V (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J Biomol NMR 56:291–301. doi:10.1007/s10858-013-9761-7
Orekhov VY, Ibraghimov IV, Billeter M (2001) MUNIN: a new approach to multi-dimensional NMR spectra interpretation. J Biomol NMR 20:49–60
Pervushin K, Eletsky A (2003) A new strategy for backbone resonance assignment in large proteins using a MQ-HACACO experiment. J Biomol NMR 25:147–152
Rezaei-Ghaleh N, Blackledge M, Zweckstetter M (2012) Intrinsically disordered proteins: from sequence and conformational properties toward drug discovery. Chembiochem 13:930–950. doi:10.1002/cbic.201200093
Shen Y, Atreya HS, Liu GH, Szyperski T (2005) G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J Am Chem Soc 127:9085–9099. doi:10.1021/Ja0501870
Shimba N et al (2004) Optimization of 13C direct detection NMR methods. J Biomol NMR 30:175–179. doi:10.1023/B:JNMR.0000048855.35771.11
Skora L, Becker S, Zweckstetter M (2010) Molten globule precursor states are conformationally correlated to amyloid fibrils of human beta-2-microglobulin. J Am Chem Soc 132:9223–9225. doi:10.1021/ja100453e
Szyperski T, Atreya HS (2006) Principles and applications of GFT projection NMR spectroscopy. Magn Reson Chem 44:51–60. doi:10.1002/mrc.1817
Takeuchi K, Frueh DP, Hyberts SG, Sun ZY, Wagner G (2010) High-resolution 3D CANCA NMR experiments for complete mainchain assignments using Cα direct detection. J Am Chem Soc 132:2945–2951. doi:10.1021/ja907717b
Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533
Uversky VN (2002) Natively unfolded proteins: a point where biology waits for physics. Protein Sci 11:739–756. doi:10.1110/ps.4210102
Uversky VN (2011) Flexible nets of malleable guardians: intrinsically disordered chaperones in neurodegenerative diseases. Chem Rev 111:1134–1166. doi:10.1021/cr100186d
Uversky VN, Oldfield CJ, Dunker AK (2008) Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 37:215–246. doi:10.1146/annurev.biophys.37.032807.125924
Wang AC, Grzesiek S, Tschudin R, Lodi PJ, Bax A (1995) Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J Biomol NMR 5:376–382
Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ (1999) Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38:16424–16431. doi:10.1021/Bi991765q
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293:321–331. doi:10.1006/jmbi.1999.3110
Zawadzka-Kazimierczuk A, Kozminski W, Billeter M (2012a) TSAR: a program for automatic resonance assignment using 2D cross-sections of high dimensionality, high-resolution spectra. J Biomol NMR 54:81–95. doi:10.1007/s10858-012-9652-3
Zawadzka-Kazimierczuk A, Kozminski W, Sanderova H, Krasny L (2012b) High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins. J Biomol NMR 52:329–337. doi:10.1007/s10858-012-9613-x
Acknowledgments
We thank Eckhard Mandelkow and Jacek Biernat for the htau23 sample. This work was in part supported by the DFG through ZW71/3-2 and ZW71/7-1.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yao, X., Becker, S. & Zweckstetter, M. A six-dimensional alpha proton detection-based APSY experiment for backbone assignment of intrinsically disordered proteins. J Biomol NMR 60, 231–240 (2014). https://doi.org/10.1007/s10858-014-9872-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-014-9872-9