Abstract
Two novel six- and seven-dimensional NMR experiments are proposed. The new experiments employ non-uniform sampling that enables achieving high resolution in four indirectly detected dimensions and synchronous sampling in the additional dimensions using projection spectroscopy principle. The resulted data sets could be processed as five-dimensional data using existing software. The experiments facilitate resonance assignment of intrinsically disordered proteins. The novel experiments were successfully tested using 1 mM sample of α-synuclein on 600 and 800 MHz NMR spectrometers equipped with standard room temperature probes. The experiments allowed backbone assignment from a 1-day acquisition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, intrinsically disordered proteins (IDPs) have attracted widespread interest in molecular biology research (Wright and Dyson 1999; Habchi et al. 2014). Their properties like structural propensity, dynamics and interactions could be effectively studied by solution NMR spectroscopy methods. However, the intrinsic disorder results in a fast conformational dynamics which causes an effective averaging of chemical shifts. Thus, very poor peak separation makes resonance assignment difficult, even for a relatively small disordered protein fragments. As a consequence, the signal overlap in the spectra strongly limits the possibility of standard NMR experiments to provide sufficient resolution for IDPs characterization. On the other hand, the fast local dynamics significantly decreases transverse relaxation rates, enabling the application of long multidimensional pulse sequences, and making possible to achieve long evolution times in order to increase the resolution and facilitate assignment of resonances.
The projection NMR spectroscopy methods are founded on the idea of Accordion Spectroscopy (Bodenhausen and Ernst 1981), and employ synchronous incrementing of two or more evolution periods in the pulse sequence. The particularly important approach of this type is simultaneous sampling of more than one chemical shift evolution, which is referred to as radial sampling. Such an option is utilized in projection spectroscopy (Kupče and Freeman 2005; Coggins et al. 2004, 2010), and requires the algebraic decoding of peak frequencies (Kim and Szyperski 2003; Koźmiński and Zhukov 2003; Hiller et al. 2005), or the reconstruction of multidimensional spectrum (Kupče and Freeman 2003).
To date, several high dimensional strategies have been proposed for backbone resonance assignment of IDPs (Reddy and Hosur 2014; Goradia et al. 2015; Yoshimura et al. 2015), including direct 13C detection (Bermel et al. 2006, 2009; Nováček et al. 2011; Pantoja-Uceda and Santoro 2013, 2014) and HA detection (Mäntylahti et al. 2010, 2011; Yao et al. 2014) which allow for revealing of proline residues. The number of dimensions, i.e. number of simultaneously correlated frequencies, could be increased by employing automated projection spectroscopy (APSY) (Hiller et al. 2007; Narayanan et al. 2010; Yao et al. 2014), or the non-uniformly sampled (NUS) high-dimensional experiments (Kazimierczuk et al. 2010; Zawadzka-Kazimierczuk et al. 2012).
APSY approach enabled the acquisition of experiments with dimensionality up to 7, by recording a set of 2D projections of high dimensionality spectrum at different angles. The high-dimensional peak list is achieved by algebraic reconstruction from automatically collected peak coordinates in the projections. The possible disadvantage of this technique is severe peak overlap on 2D spectral projections, which may influence the precision of frequency determination.
The advantage of non-uniformly sampled high-dimensionality experiments, followed by SMFT (sparse multidimensional Fourier transform) processing (Kazimierczuk et al. 2009), is the ability for visual inspection of the spectrum of the full dimensionality. Such examination of 2D cross-sections helps to assign peaks suffering from partial overlap.
SMFT processing procedure with the output in the form of a series of 2D cross-sections, requires, however, a so-called base spectrum of dimensionality of N-2, which becomes impractical for N > 5. Therefore, we propose here new 6 and 7D experiments that feature high resolution and high dimensionality resulting from the use of non-uniform sampling in the indirectly detected dimensions combined with projection spectroscopy principle. The experiments utilize NUS in four indirectly sampled dimensions, while additional frequencies are added by synchronous incrementing with t 1. Thus, the previously described 5D SMFT processing with a 3D base spectrum could be used for the proposed experiments, resulting in a 5D spectrum with additional dimensions projected into F 1.
Since the N and CO chemical shifts were found to be best dispersed in IDPs (Piai et al. 2014; Nowakowski et al. 2015), the proposed experiments are derived from 5D (H)NCO(NCA)CONH (Zawadzka-Kazimierczuk et al. 2012), by adding HN (6D) or HN and CA (7D) chemical shifts. The experimental verification was performed on a 1 mM sample of α-synuclein, employing 600 and 800 MHz NMR spectrometers with standard RT triple resonance probes.
Materials and methods
Pulse sequence
The proposed pulse sequence is based on 5D (H)NCO(NCA)CONH presented in (Zawadzka-Kazimierczuk et al. 2012) and is depicted in the Fig. 1.
HNCO(N)CACONH experiment. Top scheme shows the coherence pathway involved in the experiment. Bottom is the pulse sequence scheme. Rectangles represent hard pulses. Filled and empty symbols represent 90° and 180° pulses, respectively. 1H and 15N composite pulse decouplings are performed with WALTZ-16 (Shaka et al. 1983), at γB1/2π of 5.4 and 1.14 kHz at the 800 MHz spectrometer, and 4.0 and 0.97 kHz at 600 MHz, respectively. Simultaneous inversion of CA and CO spins was archived using 6-element composite pulse (Shaka 1985). Selective CA and CO 13C 90° (180°) pulses were applied with rf field strength adjusted to |ΔΩCA-CO|/√15 (√3). At the 800 MHz spectrometer 90° and 180°, rectangular and sinc-shaped pulses (bell-shaped at the scheme) of the duration of 40.1 (35.9) μs, and 65.8 (58.8) μs, respectively, were used. Whereas, at the 600 MHz spectrometer of, 53.5 (47.9) μs, and 87.8 (78.4) μs, respectively. Off-resonance pulses were applied using phase modulation of the carrier. The amplitude (Tm−1) of G1–G9 PFG pulses were set to: 0.212, 0.154, 0.137, 0.0926, 0.0820, 0.347, 0.154, 0.0579, 0.352 at the 800 MHz spectrometer and at 600 MHz: 0.222, 0.162, 0.143, 0.0970, 0.0858, 0.364, 0.162, 0.0606, 0.368, respectively. The PFG duration of G1–G5, G8 of 0.5 ms, G6 and G7 of 2.0 ms and G9 of 0.2 ms, were used. Evolutions for H, N, CA were in semi-constant-time mode: a i = (t i + Δ)/2; b i = t i (1 − Δ/t imax)/2; c i = Δ(1 − t i /t imax)/2 or in constant-time mode: a i = (t i + Δ)/2; b i = 0; c i = (Δ − t i )/2 where Δ stands for ΔN–H, ΔN–CO, ΔN–CA. Evolution for CO in t 2 is in real-time mode. Delays were set as follows: ΔN–H = 5.4 ms, ΔN–CO = 28 ms, ΔCO–N–CA = 28 ms, ΔN–CA = 54 ms, ΔCO–CA = 9.1 ms. The four step phase cycle was used: ϕ1 = x, -x; ϕ2 = 2x, 2(-x) and ϕrec = x, 2(-x), x = ϕ1 + ϕ2. In t 1, t 2, t 3, t 4 dimensions quadrature was accomplished using States-TPPI method, by incrementing ϕ1, ϕ2, ϕ3, ϕ4 phases, respectively. In t 5, t 6 dimensions quadrature was accomplished using States method by incrementing ϕ5, ϕ6 phases and adding additional increments to phase ϕ1 = ϕ1 + ϕ5 + ϕ6, thus, the ϕ1 phase was incremented by additional 90° for each sine modulation in t 1, t 5 and t 6. The phase ψ = x was inverted simultaneously with the last gradient (G9) pulse to achieve echo-antiecho coherence transfer selection in the indirect dimension. The coherence selection gradients (G6 and G9) were applied at magic angle (600 MHz) or along z-axis (800 MHz). 90° and 180° water 1.2 ms sinc-shaped flipback pulses were used for ϕ5 phase equal to x and y, respectively. For 5D HNCO(N)CACONH experiment t 5, t 6 and ϕ5, ϕ6 were set to 0 s and x, respectively. Additional dimensions are achieved by setting evolution times (t 5 and/or t 6) proportional to t 1. The ratio of maximum evolution times determined the respective projection angle. Note, that two different 6D and one 7D experiments could be acquired according to presented scheme
The original pulse sequence was designed to obtain both sequential and auto-correlation peaks by setting ΔN–CA delay to 28.6 ms. It surely allows easier visual inspection of the spectra. On the other hand, the doubled number of peaks increases sampling artefacts level. Moreover, lack of auto-correlation peaks present in the spectrum reduces spectral overlap, which may affect accuracy of measured peaks positions. This is especially important while studying IDPs. The coherence transfer amplitudes are described by Eqs. (1) and (2):
Assuming \({}_{{}}^{1} J_{NCA} ,{}_{{}}^{2} J_{NCA}\), \({}_{{}}^{1} J_{CACB}\) values of 11, 7 and 35 Hz (Sattler et al. 1999), respectively, the optimal choice of ΔN–CA delay length for both high intensity of sequential signal (Ni−1(ω1), COi−2(ω2), COi−1(ω3), Ni(ω4), H Ni (ω5)) and the suppression of auto-correlation signal (Ni(ω1), COi−1(ω2), COi−1(ω3), Ni(ω4), H Ni (ω5)) can be obtained with the value of 54 ms (see Fig. 2), similar approach was also presented by Nováček et al. 2011. In the present work the auto-correlation peaks suppressed version was used.
Quadrature for t 1 dimensions is achieved using a standard States-TPPI procedure. Quadratures in coevolved dimensions require recording and storing of two data sets, cosine and sine modulated, respectively. It is obtained by 90 degree shift of phases ϕ5, and ϕ5, ϕ6 (for 6D and 7D versions respectively) (Koźmiński and Zhukov 2003; Kim and Szyperski 2003). To allow standard SMFT data processing the π/2 phase shifts for sine modulated components are compensated by simultaneous π/2 shifting of ϕ1 phase, for t 1 dimension. Therefore, such procedure of processing of spectra after SMFT approach is limited to a simple co-addition to obtain spectra differing in frequency signs in coevolved dimensions.
Data processing and inspection
The presented experiments are, in fact, projections of six- and seven-dimensional experiments to the five- dimensional frequency space. Therefore, both of the presented experiments were processed as a usual 5D spectrum using SMFT approach. Last three dimensions (CO, N, HN) were fixed with chemical shifts obtained from 3D HNCO experiment. This lead to a set of 2D cross-sections showing correlations of coevolved dimensions (t 1, t 5, t 6) with CO (t 2). A simple co-addition of differently modulated data sets result with two data sets (in a case of 6D), which differ with signs of involved frequencies (7D version results in four different combinations), this procedure is schematically illustrated in the Figs. 3 and 4.
Pictorial representation of resulting peak pattern when two dimensions are coevolved. Two modulations lead to spectra I and II containing 2 peaks each. Sum and difference of obtained spectra gives two different spectra containing only one peak each, for which frequencies are a linear combinations of frequencies from coevolved dimensions. a coefficient is equal to t 2/t 1. Empty and filled circles represent positive and negative signal intensities, respectively
Pictorial representation of resulting peak pattern when three dimensions are coevolved. Four modulations lead to spectra I, II, III and IV containing 4 peaks each. Co-addition of obtained spectra gives four different spectra containing only one peak each, for which frequencies are a linear combinations of frequencies from coevolved dimensions. a, and b coefficients are equal to t 2/t 1, and t 3/t 1, respectively. Empty and filled circles represent positive and negative signal intensities, respectively
All 5D spectra were processed using ToASTD (Kazimierczuk et al. 2006) and reduced (Kazimierczuk et al. 2009) programs which take peak list from 3D HNCO as the input for SMFT routine. For 3D HNCO sampling artefacts were removed from the spectra using Signal Separation Algorithm (Stanek and Koźmiński 2010), no such procedure was performed for 5D spectra. All used programs are available at http://nmr.cent3.uw.edu.pl/software. After processing, all spectra were inspected using Sparky (Goddard and Kneller 2000).
Four distinct peaks frequencies in the projected dimension: \(\Omega_{1}\), …, \(\Omega_{4}\) on the transformations for 6D experiment are given by the Eq. (3):
In the case of 7D data the analogous expressions for peak frequencies: \(\Omega_{1}\), …, \(\Omega_{8}\) are given by the Eq. (4)
\(\Omega_{N}\),\(\Omega_{H}\), \(\Omega_{CA}\) are the values of resonance frequencies of N, H and CA nuclei of the \(n - 1\) residue with respect to the corresponding carrier offsets. \(t_{1}\), \(t_{5}\), \(t_{6}\) are maximum evolution times in N, H and CA dimensions, respectively. As a result, for 6D spectrum, a system of four equations (two of them are linearly independent) with two unknown is obtained. In the case of 7D spectrum system of 8 equations (four linearly independent) with three unknown is created. Solving these systems of equations results in \(\Omega_{N}\), \(\Omega_{H}\), \(\Omega_{CA}\) frequencies. The first system of equations (Eq. 3) is unambiguously defined, however, the second one (Eq. 4) is overdetermined. Three equations are sufficient to solve it but in order to increase the precision of a result, all four equations with equal weights were always used.
NMR spectroscopy
NMR sample contained 1 mM of 13C, 15N-labeled α-synuclein in 20 mM sodium phosphate buffer, pH 6.5, 200 mM NaCl. NMR spectra were recorded at 288 K on Agilent 800 MHz and 600 MHz spectrometers both equipped with room temperature probes. 7D HNCO(N)CACONH was acquired on the 800 MHz spectrometer in 31 h with evolution times set to 20, 30, 25, 52, 25, 30 ms for H Ni , Ni, COi−1, CAi−1, COi, Ni+1 dimensions, respectively. 6D HNCO(NCA)CONH was acquired on both, 800 MHz and 600 MHz, spectrometers in 13 and 23 h, respectively. Evolution times for the measurement on the 800 MHz spectrometer were set to: 20, 30, 25, 25, 30 ms, for H Ni , Ni, COi−1, COi, Ni+1 dimensions, respectively, and to sustain high enough resolution on the lower field evolution times were set to: 20, 40, 40, 40, 40 ms, respectively on the 600 MHz spectrometer.
Results
In order to verify results obtained from 6D and 7D projection experiments full backbone assignment of the α-synuclein was performed using 5D HN(CA)CONH (Kazimierczuk et al. 2010), 5D (H)NCO(NCA)CONH (Zawadzka-Kazimierczuk et al. 2012) and 5D HabCabCONH (Kazimierczuk et al. 2010) experiments. To avoid any discrepancies, exactly the same sample as for 6D and 7D measurements was used.
Analysis of both recorded 6D spectra led to identification of all but two expected sequential peaks. Two missing residues were the first two residues in the protein sequence which suggest that those signals were broadened beyond detection limit due to amide proton exchange. Similar result was obtained from 7D spectrum, the same set of sequential signals was identified.
Performing sequential assignment of α-synuclein due to its disordered nature can be regarded as a challenging task. Despite α-synuclein’s moderate size (140 a.a.), full potential of the “HNCO-HNCO” strategy can be seen, for example, in the case of G36CO-V37N and G73CO-V74N signals which are hardly distinguishable using COi−1Ni connectivity (see Fig. 5). Application of 6D experiment quickly resolves such ambiguity without using any additional experiment as the HN chemical shifts of aforementioned residues vary by almost 0.2 ppm (see Fig. 6). Moreover, peak positions in the resulting spectra are further differentiated when seventh dimension (CAi−1) is introduced (see Fig. 7). In addition to a better signal dispersion CA chemical shift provide partial information about residue type.
Two 2D cross-sections from 5D (H)NCO(NCA)CONH experiment (on the left) corresponding to HNCO peak positions of G36CO-V37N and G73CO-V74N signals showing COi−1Ni sequential peaks. 2D NCO projection from 3D HNCO (on the right) showing highly crowded glycine region. Unambiguous assignment is prevented due to signal overlap
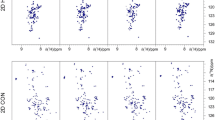
Four 2D cross-sections from 6D HNCO(NCA)CONH experiment (at the top) (++ and +− corresponds to the signs in the co-addition of obtained raw data sets) corresponding to the HNCO peak positions of G36CO-V37N and G73CO-V74N signals showing COi−1(N i H N i ) sequential peaks. Small difference in frequencies in the measured peak positions encodes additional information of HN chemical shifts. Two slices from 3D HNCO spectra obtained at HN the positions calculated from 6D experiment data (at the bottom). Unambiguous assignment is possible owing to the difference in HN chemical shifts of V37 and V74. Note that y-axis at the cross-sections from the 6D HNCO(NCA)CONH spectrum is labelled with 15N chemical shift scale, however, the peak frequencies in this dimension are given by the Eq. (3)
Eight 2D cross-sections from 7D HNCO(N)CACONH experiment (++++ , ++−, +−+− and +− + correspond to the signs in the co-addition of obtained raw data sets) corresponding to the HNCO peak positions of G36CO-V37N and G73CO-V74N signals showing CA i−1COi−1(N i H N i ) sequential peaks. While coevolution of HN leads to a more robust connectivity adding of CA frequency further differentiate signal positions due to the difference in CA chemical shifts of V37 and V74. Note that y-axis at the cross-sections from the 7D HNCO(N)CACONH spectrum is labelled with 15N chemical shift scale, however, the peak frequencies in this dimension are given by the Eq. (4)
Discussion
The most important advantage of presented approach is the establishment of highly robust connectivities due to the expansion of sequential peaks frequencies by addition of H Ni−1 frequency. Similar approach was already proposed in the literature which involved the combination of two different 5D experiments (Kazimierczuk et al. 2013; Piai et al. 2014) having shared sequential nitrogen dimension. Here it was presented that such “HNCO-HNCO” strategy can be successfully implemented within one experiment. Additional expansion to 7D experiment giving CA chemical shift can further limit number of overlapping signals as well as provide partial information needed for identifying amino acid type.
True benefit of proposed approach is the possibility of a visual inspection of resulting spectra which allows to use the expertise and the experience of a spectroscopist in resolving most difficult, severely overlapped cases. As a result, all of non-proline residues of α-synuclein were successfully identified and assigned from a single experiment (except aforementioned first two residues).
Surprisingly, even example of a moderate size α-synuclein already justifies expanding “CON-CON” strategy. Presented approach can be especially beneficial in performing the resonance assignment of proteins of a larger size than α-synuclein, since the spectra in such a case will be more crowded. What is more, sensitivity of presented experiments is high enough to record good quality spectra in a relatively short time compared to other six- and seven-dimensional approaches (Hiller et al. 2007; Yao et al. 2014), even without the use of cryogenically cooled probe.
We have demonstrated the principles of the new experiment and data inspection protocol. Although, it could be modified in several ways. The proposed experiment is fully compatible with BEST approach (Lescop et al. 2007; Solyom et al. 2013) and co-solute paramagnetic relaxation enhancement (Oktaviani et al. 2015), which could be used for acceleration of the signal repetition rate, and thus increase the number of data points acquired in a given time. Amide hydrogen exchange process, particularly effective in the IDP molecules, may cause a severe signal loss at conditions close to physiological (temperature and pH). Therefore, the pulse sequence could be modified using the aliphatic proton excitation combined with the 13C or HA detection (Mäntylahti et al. 2010). Moreover, aliphatic protons or 13C detection would allow observation of additional cross-peaks from proline residues (Bermel et al. 2006; Hellman et al. 2014). Additionally, the amide protons exchange causes further decay of 15N–1H antiphase coherences. Hence, the refocusing of 15N–1H couplings and 1H composite pulse decoupling applied during the main part of pulse sequence is used to maximize the sensitivity.
Conclusions
We presented six- and seven-dimensional experiments for backbone assignment of intrinsically disordered proteins. Described techniques combine projection spectroscopy and SMFT data processing that allow access to full dimensional spectra information. 6D experiment correlates H Ni , Ni, COi−1, H Ni−1 , Ni−1, COi−2. 7D adds additional correlation with CAi−1 dimension. We proved that these experiments can be successfully run on a standard room-temperature probe, even on a medium field NMR spectrometer (600 MHz).
References
Bermel W, Bertini I, Felli IC, Lee Y, Luchinat C, Pierattelli R (2006) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc 128:3918–3919. doi:10.1021/ja0582206
Bermel W, Bertini I, Felli IC, Pierattelli R (2009) Speeding up 13C direct detection biomolecular NMR spectroscopy. J Am Chem Soc 131:15339–15345. doi:10.1021/ja9058525
Bodenhausen G, Ernst RR (1981) The accordion experiment, a simple approach to three-dimensional NMR spectroscopy. J Magn Reson 45:367–373. doi:10.1016/0022-2364(81)90137-2
Coggins BE, Venters RA, Zhou P (2004) Generalized reconstruction of n-D NMR spectra from multiple projections: application to the 5-D HACACONH spectrum of protein G B1 domain. J Am Chem Soc 126:1000–1001. doi:10.1021/ja039430q
Coggins BE, Venters RA, Zhou P (2010) Radial sampling for fast NMR: concepts and practices over three decades. Prog Nucl Magn Reson Spec 57:381–419. doi:10.1016/j.pnmrs.2010.07.001
Goddard TD, Kneller DG (2000) SPARKY 3. University of California, San Francisco
Goradia N, Wiedemann C, Herbst C, Görlach M, Heinemann SH, Ohlenschläger O, Ramachandran R (2015) An approach to NMR assignment of intrinsically disordered proteins. ChemPhysChem 16:739–746. doi:10.1002/cphc.201402872
Habchi J, Tompa P, Longhi S, Uversky VN (2014) Introducing protein intrinsic disorder. Chem Rev 114:6561–6588. doi:10.1021/cr400514h
Hellman M, Piirainen H, Jaakola V, Permi P (2014) Bridge over troubled proline: assignment of intrinsically disordered proteins using (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H experiments concomitantly with HNCO and i(HCA)CO(CA)NH. J Biomol NMR 58:49–60. doi:10.1007/s10858-013-9804-0
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102:10876–10881. doi:10.1073/pnas.0504818102
Hiller S, Wasmer C, Wider G, Wüthrich K (2007) Sequence-specific resonance assignment of soluble nonglobular proteins by 7D APSY-NMR spectroscopy. J Am Chem Soc 129:10823–10828. doi:10.1021/ja072564+
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2006) Random sampling of evolution time space and Fourier transform processing. J Biomol NMR 36:157–168. doi:10.1007/s10858-006-9077-y
Kazimierczuk K, Zawadzka A, Koźmiński W (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson 197:219–228. doi:10.1016/j.jmr.2009.01.003
Kazimierczuk K, Zawadzka-Kazimierczuk A, Koźmiński W (2010) Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J Magn Reson 205(2):286–292. doi:10.1016/j.jmr.2010.05.012
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Koźmiński W (2013) High-dimensional NMR spectra for structural studies of biomolecules. ChemPhysChem 14:3015–3025. doi:10.1002/cphc.201300277
Kim S, Szyperski T (2003) GFT NMR—a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125:1385–1393. doi:10.1021/ja028197d
Koźmiński W, Zhukov I (2003) Multiple quadrature detection in reduced dimensionality experiments. J Biomol NMR 26:157–166. doi:10.1023/A:1023550224391
Kupče Ẽ, Freeman R (2003) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125:13958–13959. doi:10.1021/ja0582206
Kupče Ẽ, Freeman R (2005) Fast multidimensional NMR: radial sampling of evolution space. J Magn Reson 173:317–321. doi:10.1016/j.jmr.2004.12.004
Lescop E, Schanda P, Brutscher B (2007) A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson 187:163–169. doi:10.1016/j.jmr.2007.04.002
Mäntylahti S, Aitio O, Hellman M, Permi P (2010) HA-detected experiments for the backbone assignment of intrinsically disordered proteins. J Biomol NMR 47:171–181. doi:10.1007/s10858-010-9421-0
Mäntylahti S, Hellman M, Permi P (2011) Extension of the HA-detection based approach: (HCA)CON(CA)H and (HCA)NCO(CA)H experiments for the main-chain assignment of intrinsically disordered proteins. J Biomol NMR 49:99-109. doi:10.1007/s10858-011-9470-z
Narayanan RL, Duerr HN, Bilbow S, Biernat J, Mendelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132:11906–11907
Nováček J, Zawadzka-Kazimierczuk A, Papoušková V, Žídek L, Šanderová H, Krásný L, Koźmiński W, Sklenář V (2011) 15D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR 50:1–11. doi:10.1007/s10858-011-9496-2
Nowakowski M, Saxena S, Stanek J, Żerko S, Koźmiński W (2015) Applications of high dimensionality experiments to biomolecular NMR. Prog Nucl Magn Reson Spec 34:93–158. doi:10.1016/j.pnmrs.2015.07.001
Oktaviani NA, Risør MW, Lee Y, Megens RP, de Jong DH, Otten R, Scheek RM, Enghild JJ, Nielsen NC, Ikegami T, Mulder FAA (2015) Optimized co-solute paramagnetic relaxation enhancement for the rapid NMR analysis of a highly fibrillogenic peptide. J Biomol NMR 62:129–142. doi:10.1007/s10858-015-9925-8
Pantoja-Uceda D, Santoro J (2013) Direct correlation of consecutive C′–N groups in proteins: a method for the assignment of intrinsically disordered proteins. J Biomol NMR 57:57–63. doi:10.1007/s10858-013-9765-3
Pantoja-Uceda D, Santoro J (2014) New 13C-detected experiments for the assignment of intrinsically disordered proteins. J Biomol NMR 59:43–50. doi:10.1007/s10858-014-9827-1
Piai A, Hošek T, Gonnelli L, Zawadzka-Kazimierczuk A, Koźmiński W, Brutscher B, Bermel W, Pierattelli R, Felli IC (2014) “CON-CON” assignment strategy for highly flexible intrinsically disordered proteins. J Biomol NMR 60:209–218. doi:10.1007/s10858-014-9867-6
Reddy JG, Hosur RV (2014) A reduced dimensionality NMR pulse sequence and an efficient protocol for unambiguous assignment in intrinsically disordered proteins. J Biomol NMR 59:199–210. doi:10.1007/s10858-014-9839-x
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spec 34:93–158. doi:10.1016/S0079-6565(98)00025-9
Shaka AJ (1985) Composite pulses for ultra-broadband spin inversion. Chem Phys Lett 120:201–205. doi:10.1016/0009-2614(85)87040-8
Shaka AJ, Keeler J, Freeman R (1983) Evaluation of a new broadband decoupling sequence: WALTZ-16. J Magn Reson 53:313–340. doi:10.1016/0022-2364(83)90035-5
Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D, Brutscher B (2013) BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR 55:311–321. doi:10.1007/s10858-013-9715-0
Stanek J, Koźmiński W (2010) Iterative algorithm of discrete fourier transform for processing randomly sampled NMR data sets. J Biomol NMR 47:65–77. doi:10.1007/s10858-010-9411-2
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J Mol Biol 293:321–331. doi:10.1006/jmbi.1999.3110
Yao X, Becker S, Zweckstetter M (2014) A six-dimensional alpha proton detection-based APSY experiment for backbone assignment of intrinsically disordered proteins. J Biomol NMR 60:231–240. doi:10.1007/s10858-014-9872-9
Yoshimura Y, Kulminskaya NV, Mulder FAA (2015) Easy and unambiguous sequential assignments of intrinsically disordered proteins by correlating the backbone 15N or 13C′ chemical shifts of multiple contiguous residues in highly resolved 3D spectra. J Biomol NMR 61:109–121. doi:10.1007/s10858-014-9890-7
Zawadzka-Kazimierczuk A, Koźmiński W, Šanderová H, Krásný L (2012) High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins. J Biomol NMR 52:329–337. doi:10.1007/s10858-012-9613-x
Acknowledgments
This work was supported by Polish National Science Centre with PRELUDIUM grant No. 2013/11/N/ST4/01832. The study was carried out at the Biological and Chemical Research Centre, University of Warsaw, established within the project co-financed by European Union from the European Regional Development Fund under the Operational Programme Innovative Economy, 2007–2013.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Żerko, S., Koźmiński, W. Six- and seven-dimensional experiments by combination of sparse random sampling and projection spectroscopy dedicated for backbone resonance assignment of intrinsically disordered proteins. J Biomol NMR 63, 283–290 (2015). https://doi.org/10.1007/s10858-015-9987-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9987-7