Abstract
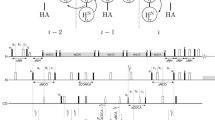
We present three novel exclusively heteronuclear 5D 13C direct-detected NMR experiments, namely (HN-flipN)CONCACON, (HCA)CONCACON and (H)CACON(CA)CON, designed for easy sequence-specific resonance assignment of intrinsically disordered proteins (IDPs). The experiments proposed have been optimized to overcome the drawbacks which may dramatically complicate the characterization of IDPs by NMR, namely the small dispersion of chemical shifts and the fast exchange of the amide protons with the solvent. A fast and reliable automatic assignment of α-synuclein chemical shifts was obtained with the Tool for SMFT-based Assignment of Resonances (TSAR) program based on the information provided by these experiments.




Similar content being viewed by others
References
Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26:131–138
Bermel W, Bertini I, Felli IC, Kümmerle R, Pierattelli R (2006a) Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J Magn Reson 178:56–64
Bermel W, Bertini I, Felli IC, Lee Y-M, Luchinat C, Pierattelli R (2006b) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc 128:3918–3919
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006c) 13C-detected protonless NMR spectroscopy of proteins in solution. Progr NMR Spectrosc 48:25–45
Bermel W, Felli IC, Kümmerle R, Pierattelli R (2008) 13C direct-detection biomolecular NMR. Concepts Magn Reson 32A:183–200
Bermel W, Bertini I, Csizmok V, Felli IC, Pierattelli R, Tompa P (2009a) H-start for exclusively heteronuclear NMR spectroscopy: the case of intrinsically disordered proteins. J Magn Reson 198:275–281
Bermel W, Bertini I, Felli IC, Pierattelli R (2009b) Speeding up 13C direct detection Biomolecular NMR experiments. J Am Chem Soc 131:15339–15345
Bermel W, Bertini I, Chill JH, Felli IC, Kumar VMV, Haba N, Pierattelli R (2012a) Aminoacid-types selective 13C detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDPs). ChemBioChem 13:2425–2432
Bermel W, Bertini I, Gonnelli L, Felli IC, Kozminski W, Piai A, Pierattelli R, Stanek J (2012b) Speeding up sequence specific assignment of IDPs. J Biomol NMR 53:293–301
Bermel W, Bruix M, Felli IC, Kumar VMV, Pierattelli R, Serrano S (2013) Improving the chemical shift dispersion of multidimensional NMR spectra of intrinsically disordered proteins. J Biomol NMR 55:231–237
Böhlen J-M, Bodenhausen G (1993) Experimental aspects of chirp NMR spectroscopy. J Magn Reson Ser A 102:293–301
Csizmok V, Felli IC, Tompa P, Banci L, Bertini I (2008) Structural and dynamic characterization of intrinsically disordered human securin by NMR. J Am Chem Soc 130:16873–16879
Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J Biomol NMR 6:277–293
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC (2001) Intrinsically disordered protein. J Mol Graph Model 19:26–59
Dyson HJ, Wright PE (2001) Nuclear magnetic resonance methods for the elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–271
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Emsley L, Bodenhausen G (1992) Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J Magn Reson 97:135–148
Gil S, Hosek T, Solyom Z, Kümmerle R, Brutscher B, Pierattelli R, Felli IC (2013) NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew Chem Int Ed. doi:10.1002/anie.201304272
Goddard TD, Kneller DG (2000) SPARKY 3. University of California, San Francisco
Haba NY, Gross R, Novacek J, Shaked H, Zidek L, Barda-Saad M, Chill JH (2013) NMR determines transient structure and dynamics in the disordered C-terminal domain of WASp interacting protein. Biophysical J 105:481–493
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102:10876–10881
Hsu ST, Bertoncini CW, Dobson CM (2009) Use of protonless NMR spectroscopy to alleviate the loss of information resulting from exchange-broadening. J Am Chem Soc 131:7222–7223
Huang C, Ren G, Zhou H, Wang C (2005) A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr Purif 42:173–177
Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I (2006) Random sampling of evolution time space and Fourier transform processing. J Biomol NMR 36:157–168
Kazimierczuk K, Zawadzka A, Kozminski W (2008) Optimization of random time domain sampling in multidimensional NMR. J Magn Reson 192:123–130
Kazimierczuk K, Zawadzka A, Kozminski W (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson 197:219–228
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W (2010) Random sampling in multidimensional NMR spectroscopy. Prog NMR Spectrosc 57:420–434
Knoblich K, Whittaker S, Ludwig C, Michiels P, Jiang T, Schafflhausen B, Günther U (2009) Backbone assignment of the N-terminal polyomavirus large T antigen. Biomol NMR Assign 3:119–123
Kumar D, Hosur RV (2011) hNCOcanH pulse sequence and a robust protocol for rapid and unambiguous assignment of backbone ((1) H(N), (15) N and (13) C’) resonances in (15) N/(13) C-labeled proteins. Magn Reson Chem 49:575–583
Mantylahti S, Hellman M, Permi P (2011) Extension of the HA-detection based approach: (HCA)CON(CA)H and (HCA)NCO(CA)H experiments for the main-chain assignment of intrinsically disordered proteins. J Biomol NMR 49:99–109
Mittag T, Forman-Kay J (2007) Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol 17:3–14
Narayanan RL, Duerr HN, Bilbow S, Biernat J, Mendelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132:11906–11907
Novacek J, Zawadzka-Kazimierczuk A, Papoušková V, Zidek L, Sanderová H, Krasny L, Kozminski W, Sklenar V (2011) 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR 50:1–11
Novacek J, Haba NY, Chill JH, Zidek L, Sklenar V (2012) 4D Non-uniformly sampled HCBCACON and (1) J(NC (α))-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J Biomol NMR 53:139–148
Novacek J, Janda L, Dopitova R, Zidek L, Sklenar V (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J Biomol NMR 56:291–301
O’Hare B, Benesi AJ, Showalter SA (2009) Incorporating 1H chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J Magn Reson 200:354–358
Panchal SC, Bhavesh NS, Hosur RV (2001) Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations (13C, 15N) labeled proteins: application to unfolded proteins. J Biomol NMR 20:135–147
Pantoja-Uceda D, Santoro J (2012) New amino acid residue type identification experiments valid for protonated and deuterated proteins. J Biomol NMR 54:145–153
Pantoja-Uceda D, Santoro J (2013) Direct correlation of consecutive C’–N groups in proteins: a method for the assignment of intrinsically disordered proteins. J Biomol NMR 57:57–63
Pérez Y, Gairi M, Pons M, Bernadò P (2009) Structural characterization of the natively unfolded N-terminal domain of human c-Src kinase: insights into the role of phosphorylation of the unique domain. J Mol Biol 391:136–148
Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK (2007) Intrinsic disorder and functional proteomics. Biophys J 92:1439–1456
Schanda P, Forge V, Brutscher B (2007) Protein folding and unfolding studied at atomic resolution by fast two-dimensional NMR spectroscopy. Proc Natl Acad Sci USA 104:11257–11262
Shaka AJ, Keeler J, Freeman R (1983) Evaluation of a new broadband decoupling sequence: WALTZ-16. J Magn Reson 53:313–340
Shaka AJ, Barker PB, Freeman R (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552
Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D, Brutscher B (2013) BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR 55:311–321
Thakur A, Chandra K, Dubey A, D’Silva P, Atreya HS (2013) Rapid characterization of hydrogen exchange in proteins. Angew Chem 52:2440–2443
Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533
Tompa P (2009) Structure and function of intrinsically disordered proteins. CRC Press, Boca Raton
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Shulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL (2007) BioMagResBank. Nucleic Acids Res 36:D402–D408
Uversky VN, Gillespie JR, Fink AL (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Genet 41:415–427
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293:321–331
Zawadzka-Kazimierczuk A, Kozminski W, Billeter M (2012) TSAR: a program for automatic resonance assignment using 2D cross-sections of high dimensionality, high-resolution spectra. J Biomol NMR 54:81–95
Acknowledgments
This work was supported in part by the EC 7th Framework program BioNMR (contract 261863), by the EC Marie Curie ITN program IDPbyNMR (contract 264257) and by grant number IP2012 062772, funded by Polish Ministry of Science and Higher Education for years 2013–2014. AZK thanks the Foundation for Polish Science for support with the START and the POMOST programs.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bermel, W., Felli, I.C., Gonnelli, L. et al. High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR 57, 353–361 (2013). https://doi.org/10.1007/s10858-013-9793-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9793-z