Abstract
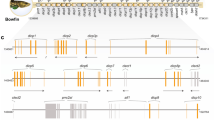
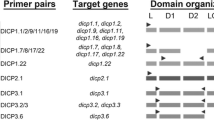
Multiple novel immunoglobulin-like transcripts (NILTs) have been identified from salmon, trout, and carp. NILTs typically encode activating or inhibitory transmembrane receptors with extracellular immunoglobulin (Ig) domains. Although predicted to provide immune recognition in ray-finned fish, we currently lack a definitive framework of NILT diversity, thereby limiting our predictions for their evolutionary origin and function. In order to better understand the diversity of NILTs and their possible roles in immune function, we identified five NILT loci in the Atlantic salmon (Salmo salar) genome, defined 86 NILT Ig domains within a 3-Mbp region of zebrafish (Danio rerio) chromosome 1, and described 41 NILT Ig domains as part of an alternative haplotype for this same genomic region. We then identified transcripts encoded by 43 different NILT genes which reflect an unprecedented diversity of Ig domain sequences and combinations for a family of non-recombining receptors within a single species. Zebrafish NILTs include a sole putative activating receptor but extensive inhibitory and secreted forms as well as membrane-bound forms with no known signaling motifs. These results reveal a higher level of genetic complexity, interindividual variation, and sequence diversity for NILTs than previously described, suggesting that this gene family likely plays multiple roles in host immunity.






Similar content being viewed by others
Data availability
New transcriptome sequence read data used in this study have been deposited to NCBI under the project accession number PRJNA672972. Custom Perl and R scripts used in the processing and formatting of data can be found on https://github.com/djwcisel/nilts.
References
Allendorf FW, Thorgaard GH (1984) Tetraploidy and the evolution of salmonid fishes. In: Turner BJ (ed) Evolutionary genetics of fishes. Springer, US, Boston, MA, pp 1–53
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Barrow AD, Trowsdale J (2006) You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol 36:1646–1653
Berthelot C, Brunet F, Chalopin D et al (2014) The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun 5:3657
Bezbradica JS, Medzhitov R (2012) Role of ITAM signaling module in signal integration. Curr Opin Immunol 24:58–66
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Braasch I, Gehrke AR, Smith JJ et al (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437
Brown KH, Dobrinski KP, Lee AS et al (2012) Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A 109:529–534
Cavanaugh JE (2016) Model Selection: Bayesian Information Criterion. Wiley StatsRef: Statistics Reference Online. https://doi.org/10.1002/9781118445112.stat00247.pub2
Clark GJ, Ju X, Tate C, Hart DNJ (2009) The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol 30:209–217
Desai S, Heffelfinger AK, Orcutt TM et al (2008) The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. BMC Evol Biol 8:177
Dirscherl H, Yoder JA (2015) A nonclassical MHC class I U lineage locus in zebrafish with a null haplotypic variant. Immunogenetics 67:501–513
Dirscherl H, Yoder JA (2014) Characterization of the Z lineage major histocompatability complex class I genes in zebrafish. Immunogenetics 66:185–198
Dornburg A, Near TJ (2021) The emerging phylogenetic perspective on the evolution of actinopterygian fishes. Annu Rev Ecol Evol Syst 52:427–452
Dornburg A, Townsend JP, Friedman M, Near TJ (2014) Phylogenetic informativeness reconciles ray-finned fish molecular divergence times. BMC Evol Biol 14
Dornburg A, Wcisel DJ, Zapfe K et al (2021) Holosteans contextualize the role of the teleost genome duplication in promoting the rise of evolutionary novelties in the ray-finned fish innate immune system. Immunogenetics 73:479–497
Dornburg A, Yoder JA (2022) On the relationship between extant innate immune receptors and the evolutionary origins of jawed vertebrate adaptive immunity. Immunogenetics 74:111–128
Eddy SR, the HMMER development team. HMMER, version 3.3. 2015. http://hmmer.org
Fernández R, Gabaldón T (2020) Gene gain and loss across the metazoan tree of life. Nat Ecol Evol 4:524–533
Flajnik MF, Tlapakova T, Criscitiello MF et al (2012) Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7’s historical relationship with the MHC. Immunogenetics 64:571–590
Gasiorowski RE, Ju X, Hart DNJ, Clark GJ (2013) CD300 molecule regulation of human dendritic cell functions. Immunol Lett 149:93–100
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haffter P, Granato M, Brand M et al (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1–36
Hamuro K, Suetake H, Saha NR et al (2007) A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J Immunol 178:5682–5689
Harpaz Y, Chothia C (1994) Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol 238:528–539
Hiby SE, Walker JJ, O’shaughnessy KM et al (2004) Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med 200:957–965
Hoang DT, Chernomor O, von Haeseler A et al (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Honjo Y, Takano K, Ichinohe T (2021) Characterization of novel zebrafish MHC class I U lineage genes and their haplotype. Dev Comp Immunol 116:103952
Howe K, Clark MD, Torroja CF et al (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503
Hwang WY, Fu Y, Reyon D et al (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Jack RS (2015) Evolution of immunity and pathogens. Results Probl Cell Differ 57:1–20
Joerink M, Ribeiro CMS, Stet RJM et al (2006) Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol 177:61–69
Kapustin Y, Souvorov A, Tatusova T, Lipman D (2008) Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct 3:20
Klesney-Tait J, Turnbull IR, Colonna M (2006) The TREM receptor family and signal integration. Nat Immunol 7:1266–1273
Kock H, Fischer U (2008) A novel immunoglobulin-like transcript from rainbow trout with two Ig-like domains and two isoforms. Mol Immunol 45:1612–1622
Kortum AN, Rodriguez-Nunez I, Yang J et al (2014) Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 66:267–279
Kruiswijk CP, Hermsen TT, Westphal AH et al (2002) A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J Immunol 169:1936–1947
Krzywinski M, Schein J, Birol I et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
LaFave MC, Varshney GK, Vemulapalli M et al (2014) A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics 198:167–170
Lanier LL (2008) Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9:495–502
Lefranc MP, Giudicelli V, Ginestoux C et al (1999) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res 27:209–212
Lefranc M-P, Pommié C, Ruiz M et al (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 27:55–77
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Lien S, Koop BF, Sandve SR et al (2016) The Atlantic salmon genome provides insights into rediploidization. Nature 533:200–205
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Marçais G, Delcher AL, Phillippy AM et al (2018) MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14:e1005944
Martin MP, Carrington M (2013) Immunogenetics of HIV disease. Immunol Rev 254:245–264
McConnell SC, Hernandez KM, Wcisel DJ et al (2016) Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A 113:E5014–E5023
McConnell SC, Hernandez KM, Andrade J et al (2022) Immune gene variation associated with chromosome-scale differences among individual zebrafish genomes. bioRxiv. https://doi.org/10.1101/2022.06.08.495387
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Misra MK, Augusto DG, Martin GM et al (2018) Report from the killer-cell immunoglobulin-like receptors (KIR) component of the 17th International HLA and Immunogenetics Workshop. Hum Immunol 79:825–833
Mizgirev IV, Revskoy S (2014) A new zebrafish model for experimental leukemia therapy. Cancer Biol Ther 9:895–902
Montgomery BC, Cortes HD, Mewes-Ares J et al (2011) Teleost IgSF immunoregulatory receptors. Dev Comp Immunol 35:1223–1237
Near TJ, Eytan RI, Dornburg A et al (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci 109:13698–13703
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Østergaard AE, Lubieniecki KP, Martin SAM et al (2010) Genomic organisation analysis of novel immunoglobulin-like transcripts in Atlantic salmon (Salmo salar) reveals a tightly clustered and multigene family. BMC Genomics 11:697
Østergaard AE, Martin SAM, Wang T et al (2009) Rainbow trout (Oncorhynchus mykiss) possess multiple novel immunoglobulin-like transcripts containing either an ITAM or ITIMs. Dev Comp Immunol 33:525–532
Panagos PG, Dobrinski KP, Chen X et al (2006) Immune-related, lectin-like receptors are differentially expressed in the myeloid and lymphoid lineages of zebrafish. Immunogenetics 58:31–40
Papkou A, Guzella T, Yang W et al (2019) The genomic basis of Red Queen dynamics during rapid reciprocal host-pathogen coevolution. Proc Natl Acad Sci U S A 116:923–928
Pleyte KA, Duncan SD, Phillips RB (1992) Evolutionary relationships of the salmonid fish genus Salvelinus inferred from DNA sequences of the first internal transcribed spacer (ITS 1) of ribosomal DNA. Mol Phylogenet Evol 1:223–230
Robinson JT, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Rodríguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA (2014) Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: deciphering the differences. Dev Comp Immunol 46:24–34
Rodriguez-Nunez I, Wcisel DJ, Litman RT et al (2016) The identification of additional zebrafish DICP genes reveals haplotype variation and linkage to MHC class I genes. Immunogenetics 68:295–312
Sheng X, Qian X, Tang X et al (2018) Polymeric immunoglobulin receptor mediates immune excretion of mucosal IgM-antigen complexes across intestinal epithelium in flounder (Paralichthys olivaceus). Front Immunol 9:1562
Sievers F, Higgins DG (2021) The Clustal Omega Multiple Alignment Package. Methods Mol Biol 2231:3–16
Stafford JL, Bengtén E, Du Pasquier L et al (2006) A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 58:758–773
Stet RJM, Hermsen T, Westphal AH et al (2005) Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics 57:77–89
Tadiso TM, Sharma A, Hordvik I (2011) Analysis of polymeric immunoglobulin receptor- and CD300-like molecules from Atlantic salmon. Mol Immunol 49:462–473
Taylor LS, Paul SP, McVicar DW (2000) Paired inhibitory and activating receptor signals. Rev Immunogenet 2:204–219
Thompson AW, Hawkins MB, Parey E et al (2021) The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat Genet 53:1373–1384
Traver D, Yoder JA (2020) Chapter 19 - immunology. In: Cartner SC, Eisen JS, Farmer SC et al (eds) The zebrafish in biomedical research. Academic Press, pp 191–216
Truett GE, Heeger P, Mynatt RL et al (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52–54
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wcisel DJ, Ota T, Litman GW, Yoder JA (2017) Spotted gar and the evolution of innate immune receptors. J Exp Zool B Mol Dev Evol 328:666–684
Wcisel DJ, Yoder JA (2016) The confounding complexity of innate immune receptors within and between teleost species. Fish Shellfish Immunol 53:24–34
Wong RY, Perrin F, Oxendine SE et al (2012) Comparing behavioral responses across multiple assays of stress and anxiety in zebrafish (Danio rerio). Behaviour 149:1205–1240
Yoder JA, Litman GW (2011) The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63:123–141
Acknowledgements
We thank Dereje Jima (North Carolina State University) for early discussions about zebrafish NILTs, James (Thomas) Howard (North Carolina State University) for technical support, and Shawn Burgess (National Human Genome Research Institute), John Godwin (North Carolina State University), David Langenau (Mass General Research Institute), John Rawls (Duke University), and Sergei Revskoy (University of Kentucky College of Medicine) for zebrafish lines.
Funding
This work was supported by the National Science Foundation (IOS1755330 to JAY and IOS1755242 to AD), the National Institutes of Health (R01 AI057559 to GWL and JAY and R01 AI23337 to GWL), the National Evolutionary Synthesis Center, NSF EF0905606 (DJW), the Triangle Center for Evolutionary Medicine (AD and JAY), and the Chicago Biomedical Consortium (CCT Searle Fund to JdJ and SCM), and by services provided by the University of Chicago Genomics Core Facility and Bioinformatics Core Facility which are supported by the UChicago Medicine Comprehensive Cancer Center NCI Cancer Support Center Grant (P30 CA014599).
Author information
Authors and Affiliations
Contributions
DJW, GWL, and JAY conceived the project; DJW assembled transcriptome, datamined public databases, and wrote custom scripts to facilitate these processes; AD performed phylogenetic analyses; SCM, KMH, JA, and JdJ completed genome sequencing and assembly; DJW, AD, and JAY collected and analyzed the data, created graphics, and wrote the manuscript; JAY supervised the project. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wcisel, D.J., Dornburg, A., McConnell, S.C. et al. A highly diverse set of novel immunoglobulin-like transcript (NILT) genes in zebrafish indicates a wide range of functions with complex relationships to mammalian receptors. Immunogenetics 75, 53–69 (2023). https://doi.org/10.1007/s00251-022-01270-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-022-01270-9