Abstract
For over half a century, deciphering the origins of the genomic loci that form the jawed vertebrate adaptive immune response has been a major topic in comparative immunogenetics. Vertebrate adaptive immunity relies on an extensive and highly diverse repertoire of tandem arrays of variable (V), diversity (D), and joining (J) gene segments that recombine to produce different immunoglobulin (Ig) and T cell receptor (TCR) genes. The current consensus is that a recombination-activating gene (RAG)-like transposon invaded an exon of an ancient innate immune VJ-bearing receptor, giving rise to the extant diversity of Ig and TCR loci across jawed vertebrates. However, a model for the evolutionary relationships between extant non-recombining innate immune receptors and the V(D)J receptors of the jawed vertebrate adaptive immune system has only recently begun to come into focus. In this review, we provide an overview of non-recombining VJ genes, including CD8β, CD79b, natural cytotoxicity receptor 3 (NCR3/NKp30), putative remnants of an antigen receptor precursor (PRARPs), and the multigene family of signal-regulatory proteins (SIRPs), that play a wide range of roles in immune function. We then focus in detail on the VJ-containing novel immune-type receptors (NITRs) from ray-finned fishes, as recent work has indicated that these genes are at least 50 million years older than originally thought. We conclude by providing a conceptual model of the evolutionary origins and phylogenetic distribution of known VJ-containing innate immune receptors, highlighting opportunities for future comparative research that are empowered by this emerging evolutionary perspective.




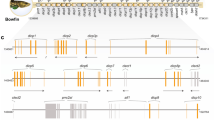
adapted from Dornburg and Near (2021). Blue rectangles indicate species from which NITRs were previously reported (Table 2). Yellow rectangles reflect species from which NITRs were identified using BLAST searches of publicly available sequences (terminal taxa). Roman numerals correspond to major lineages of Acanthomorph fishes (Dornburg and Near 2021) for reference: (I) Ophidiiformes, (II) Batrachoidiformes, (III) Gobiiformes, (IV) Syngnathiformes, (V) Blenniiformes, (VI) Carangiformes, (VII) Perciformes, (VIII) Centrachiformes, (IX) Labriformes, and (X) Acanthuriformes. Fish illustrations are provided by T. Near


Similar content being viewed by others
References
Agrawal A, Eastman QM, Schatz DG (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744–751
Alfaro ME, Santini F, Brock C et al (2009) Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci U S A 106:13410–13414
Allcock RJN, Barrow AD, Forbes S et al (2003) The human TREM gene cluster at 6p21.1 encodes both activating and inhibitory single IgV domain receptors and includes NKp44. Eur J Immunol 33:567–577
Allison JP, McIntyre BW, Bloch D (1982) Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J Immunol 129:2293–2300
Barclay AN, Brown MH (2006) The SIRP family of receptors and immune regulation. Nat Rev Immunol 6:457–464
Barrow AD, Trowsdale J (2006) You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol 36:1646–1653
Boehm T, Hirano M, Holland SJ et al (2018) Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol 36:19–42
Braasch I, Gehrke AR, Smith JJ et al (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437
Brown EJ, Frazier WA (2001) Integrin-associated protein (CD47) and its ligands. Trends Cell Biol 11:130–135
Cannon JP, Haire RN, Magis AT et al (2008) A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity 29:228–237
Castro R, Bernard D, Lefranc MP et al (2011) T cell diversity and TcR repertoires in teleost fish. Fish Shellfish Immunol 31:644–654
Chen R, Zhang L, Qi J et al (2018) Discovery and analysis of invertebrate IgVJ-C2 structure from amphioxus provides insight into the evolution of the Ig superfamily. J Immunol 200:2869–2881
Chien Y, Becker DM, Lindsten T et al (1984) A third type of murine T-cell receptor gene. Nature 312:31–35
Chothia C, Lesk AM (1987) Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 196:901–917
Cowman PF, Parravicini V, Kulbicki M, Floeter SR (2017) The biogeography of tropical reef fishes: endemism and provinciality through time. Biol Rev Camb Philos Soc 92:2112–2130
Danilova N, Amemiya CT (2009) Going adaptive: the saga of antibodies. Ann N Y Acad Sci 1168:130–155
Davis AM, Unmack PJ, Pusey BJ et al (2012) Marine-freshwater transitions are associated with the evolution of dietary diversification in terapontid grunters (Teleostei: Terapontidae). J Evol Biol 25:1163–1179
Desai S, Heffelfinger AK, Orcutt TM et al (2008) The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. BMC Evol Biol 8:177
Dong M, Fu Y, Yu C et al (2005) Identification and characterisation of a homolog of an activation gene for the recombination activating gene 1 (RAG 1) in amphioxus. Fish Shellfish Immunol 19:165–174
Dooley H, Flajnik MF (2006) Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 30:43–56
Dornburg A, Federman S, Lamb AD et al (2017) Cradles and museums of Antarctic teleost biodiversity. Nat Ecol Evol 1:1379–1384
Dornburg A, Moore J, Beaulieu JM et al (2015) The impact of shifts in marine biodiversity hotspots on patterns of range evolution: evidence from the Holocentridae (squirrelfishes and soldierfishes). Evolution 69:146–161
Dornburg A, Near TJ (2021) The emerging phylogenetic perspective on the evolution of actinopterygian fishes. Annu Rev Ecol Evol Syst 52 https://doi.org/10.1146/annurev-ecolsys-122120-122554
Dornburg A, Wang Z, Wang J, et al (2021a) Comparative genomics within and across bilaterians illuminates the evolutionary history of ALK and LTK proto-oncogene origination and diversification. Genome Biol Evol 13(1):evaa228 https://doi.org/10.1093/gbe/evaa228
Dornburg A, Wcisel DJ, Zapfe K et al (2021) Holosteans contextualize the role of the teleost genome duplication in promoting the rise of evolutionary novelties in the ray-finned fish innate immune system. Immunogenetics 73:479–497. https://doi.org/10.1007/s00251-021-01225-6
Eason DD, Cannon JP, Haire RN et al (2004) Mechanisms of antigen receptor evolution. Semin Immunol 16:215–226
Edelman GM, Gall WE (1969) The antibody problem. Annu Rev Biochem 38:415–466
Evenhuis J, Bengtén E, Snell C et al (2007) Characterization of additional novel immune type receptors in channel catfish, Ictalurus punctatus. Immunogenetics 59:661–671
Fellah JS, Tuffèry P, Etchebest C et al (2002) Cloning and modeling of CD8 beta in the amphibian ambystoma Mexicanum. Evolutionary conserved structures for interactions with major histocompatibility complex (MHC) class I molecules. Gene 288:95–102
Fernández R, Gabaldón T (2020) Gene gain and loss across the metazoan tree of life. Nat Ecol Evol 4:524–533
Ferraresso S, Kuhl H, Milan M et al (2009) Identification and characterisation of a novel immune-type receptor (NITR) gene cluster in the European sea bass, Dicentrarchus labrax, reveals recurrent gene expansion and diversification by positive selection. Immunogenetics 61:773–788
Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:438–453
Flajnik MF, Tlapakova T, Criscitiello MF et al (2012) Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7’s historical relationship with the MHC. Immunogenetics 64:571–590
Fu Y, Yang Z, Huang J, Cheng X et al (2019) Identification of two nonrearranging IgSF genes in chicken reveals a novel family of putative remnants of an antigen receptor precursor. J Immunol 202:1992–2004
Ghaffari SH, Lobb CJ (1999) Structure and genomic organization of a second cluster of immunoglobulin heavy chain gene segments in the channel catfish. J Immunol 162:1519–1529
Guselnikov SV, Najakshin AM, Taranin AV (2003) Fugu rubripes possesses genes for the entire set of the ITAM-bearing transmembrane signal subunits. Immunogenetics 55:472–479
Haruta C, Suzuki T, Kasahara M (2006) Variable domains in hagfish: NICIR is a polymorphic multigene family expressed preferentially in leukocytes and is related to lamprey TCR-like. Immunogenetics 58:216–225
Haskins K, Kappler J, Marrack P (1984) The major histocompatibility complex-restricted antigen receptor on T cells. Annu Rev Immunol 2:51–66
Hawke NA, Yoder JA, Haire RN, Mueller MG, Litman RT, Miracle AL, Stuge T, Shen L, Miller N, Litman GW (2001) Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci U S A 98:13832–13837
Hedrick SM, Cohen DI, Nielsen EA, Davis MM (1984a) Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 308:149–153
Hedrick SM, Nielsen EA, Kavaler J et al (1984b) Sequence relationships between putative T-cell receptor polypeptides and immunoglobulins. Nature 308:153–158
Hirano M, Guo P, McCurley N et al (2013) Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 501:435–438
Holland SJ, Gao M, Hirano M et al (2014) Selection of the lamprey VLRC antigen receptor repertoire. Proc Natl Acad Sci U S A 111:14834–14839
Hsu E (2018) Immune system receptors in vertebrates: immunoglobulins. Reference Module in Life Sciences. https://doi.org/10.1016/B978-0-12-809633-8.20721-8
Hsu E (2009) V(D)J recombination: of mice and sharks. Adv Exp Med Biol 650:166–179
Hsu E, Criscitiello MF (2006) Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol 177:2452–2462
Inoue J, Sato Y, Sinclair R et al (2015) Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci U S A 112:14918–14923
Isakov N (1997) ITIMs and ITAMs. The yin and yang of antigen and Fc receptor-linked signaling machinery. Immunol Res 16:85–100
Katsukura H, Murakami R, Chijiiwa Y, Otsuka A, Tanaka M, Nakashima K, Ono M (2001) Structure of the beta-chain (B29) gene of the chicken B-cell receptor and conserved collinearity with genes for potential skeletal muscle sodium channel and growth hormone. Immunogenetics 53:770–775
Khalturin K, Panzer Z, Cooper MD, Bosch TCG (2004) Recognition strategies in the innate immune system of ancestral chordates. Mol Immunol 41:1077–1087
Kinlein A, Janes ME, Kincer J et al (2021) Analysis of shark NCR3 family genes reveals primordial features of vertebrate NKp30. Immunogenetics 73:333–348
Kokubu F, Hinds K, Litman R et al (1988a) Complete structure and organization of immunoglobulin heavy chain constant region genes in a phylogenetically primitive vertebrate. EMBO J 7:1979–1988
Kokubu F, Hinds K, Litman R et al (1987) Extensive families of constant region genes in a phylogenetically primitive vertebrate indicate an additional level of immunoglobulin complexity. Proc Natl Acad Sci U S A 84:5868–5872
Kokubu F, Litman R, Shamblott MJ et al (1988b) Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J 7:3413–3422
Kruse PH, Matta J, Ugolini S, Vivier E (2014) Natural cytotoxicity receptors and their ligands. Immunol Cell Biol 92:221–229
Lanier LL (1998) NK cell receptors. Annu Rev Immunol 16:359–393
Laun K, Coggill P, Palmer S et al (2006) The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like Loci. PLoS Genet 2:e73
Lee SS, Fitch D, Flajnik MF, Hsu E (2000) Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med 191:1637–1648
Li J, Das S, Herrin BR et al (2013a) Definition of a third VLR gene in hagfish. Proc Natl Acad Sci U S A 110:15013–15018
Li R, Wang T, Bird S et al (2013b) B cell receptor accessory molecule CD79α: characterisation and expression analysis in a cartilaginous fish, the spiny dogfish (Squalus acanthias). Fish Shellfish Immunol 34:1404–1415
Litman GW, Frommel D, Finstad J, Good RA (1971a) Evolution of the immune response. X. Immunoglobulins of the bowfin: subunit nature. J Immunol 107:881–888
Litman GW, Frommel D, Finstad J, Good RA (1971b) The evolution of the immune response. IX. Immunoglobulins of the bowfin: purification and characterization. J Immunol 106:747–754
Litman GW, Hawke NA, Yoder JA (2001) Novel immune-type receptor genes. Immunol Rev 181:250–259
Litman GW, Rast JP, Fugmann SD (2010) The origins of vertebrate adaptive immunity. Nat Rev Immunol 10:543–553
Mashoof S, Criscitiello MF (2016) Fish Immunoglobulins Biology 5:45. https://doi.org/10.3390/biology5040045
Matlung HL, Szilagyi K, Barclay NA, van den Berg TK (2017) The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol Rev 276:145–164
Meng F, Wang R, Xu T (2014) Identification of 21 novel immune-type receptors in miiuy croaker and expression pattern of three typical inhibitory members. Dev Comp Immunol 45:269–277
Meuer SC, Fitzgerald KA, Hussey RE et al (1983) Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J Exp Med 157:705–719
Montgomery BC, Cortes HD, Mewes-Ares J et al (2011) Teleost IgSF immunoregulatory receptors. Dev Comp Immunol 35:1223–1237
Moretta A, Bottino C, Vitale M et al (2001) Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 19:197–223
Murphy KM, Weaver C (2016) Janeway’s immunobiology: ninth international student edition. W.W. Norton & Company
Nakatani M, Miya M, Mabuchi K et al (2011) Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol Biol 11:177
Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B (2021) Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat Commun 12:4489
Near TJ, Dornburg A, Eytan RI et al (2013) Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci U S A 110:12738–12743
Near TJ, Eytan RI, Dornburg A et al (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A 109:13698–13703
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
North B, Lehmann A, Dunbrack RL Jr (2011) A new clustering of antibody CDR loop conformations. J Mol Biol 406:228–256
Ohta Y, Goetz W, Hossain MZ et al (2006) Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol 176:3674–3685
Ohta Y, Kasahara M, O’Connor TD, Flajnik MF (2019) Inferring the “primordial immune complex”: origins of MHC class I and antigen receptors revealed by comparative genomics. J Immunol 203:1882–1896
Oronsky B, Carter C, Reid T et al (2020) Just eat it: a review of CD47 and SIRP-α antagonism. Semin Oncol 47:117–124
Ott JA, Ohta Y, Flajnik MF, Criscitiello MF (2021) Lost structural and functional inter-relationships between Ig and TCR loci in mammals revealed in sharks. Immunogenetics 73:17–33
Pancer Z, Amemiya CT, Ehrhardt GRA et al (2004a) Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430:174–180
Pancer Z, Mayer WE, Klein J, Cooper MD (2004b) Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci U S A 101:13273–13278
Pancer Z, Saha NR, Kasamatsu J et al (2005) Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci U S A 102:9224–9229
Papermaster BW, Condie RM, Finstad J, Good RA (1964) Evolution of the immune response. I. The phylogenetic development of adaptive immunologic responsiveness in vertebrates. J Exp Med 119:105–130
Pende D, Parolini S, Pessino A et al (1999) Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 190:1505–1516
Pessino A, Sivori S, Bottino C et al (1998) Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med 188:953–960
Pettinello R, Dooley H (2014) The Immunoglobulins of Cold-Blooded Vertebrates Biomolecules 4:1045–1069
Piyaviriyakul P, Kondo H, Hirono I, Aoki T (2007) A novel immune-type receptor of Japanese flounder (Paralichthys olivaceus) is expressed in both T and B lymphocytes. Fish Shellfish Immunol 22:467–476
Prum RO, Berv JS, Dornburg A et al (2015) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569–573
Rast JP, Anderson MK, Ota T et al (1994) Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics 40:83–99
Rast JP, Haire RN, Litman RT et al (1995) Identification and characterization of T-cell antigen receptor-related genes in phylogenetically diverse vertebrate species. Immunogenetics 42:204–212
Rast JP, Litman GW (1998) Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev 166:79–86
Reynaud CA, Dahan A, Anquez V, Weill JC (1989) Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell 59:171–183
Sahoo M, Edholm E-S, Stafford JL et al (2008) B cell receptor accessory molecules in the channel catfish, Ictalurus punctatus. Dev Comp Immunol 32:1385–1397
Sakaguchi N, Matsuo T, Nomura J et al (1993) Immunoglobulin receptor-associated molecules. Adv Immunol 54:337–392
Sato A, Mayer WE, Klein J (2003) A molecule bearing an immunoglobulin-like V region of the CTX subfamily in amphioxus. Immunogenetics 55:423–427
Sayegh CE, Demaries SL, Pike KA et al (2000) The chicken B-cell receptor complex and its role in avian B-cell development. Immunol Rev 175:187–200
Sigel MM, Voss EW Jr, Rudikoff S (1972) Binding properties of shark immunoglobulins. Comp Biochem Physiol A Comp Physiol 42:249–259
Simakov O, Marlétaz F, Yue JX, O’Connell B, Jenkins J, Brandt A, Calef R, Tung CH, Huang TK, Schmutz J, Satoh N, Yu JK, Putnam NH, Green RE, Rokhsar DS (2020) Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol 4:820–830
Siqueira AC, Bellwood DR, Cowman PF (2019) Historical biogeography of herbivorous coral reef fishes: the formation of an Atlantic fauna. J Biogeogr 46:1611–1624
Strong SJ, Mueller MG, Litman RT et al (1999) A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci U S A 96:15080–15085
Suetake H, Araki K, Akatsu K et al (2007) Genomic organization and expression of CD8alpha and CD8beta genes in fugu Takifugu rubripes. Fish Shellfish Immunol 23:1107–1118
Sun Y, Wei Z, Li N, Zhao Y (2013) A comparative overview of immunoglobulin genes and the generation of their diversity in tetrapods. Dev Comp Immunol 39:103–109
Sutoh Y, Kasahara M (2021) The immune system of jawless vertebrates: insights into the prototype of the adaptive immune system. Immunogenetics 73:5–16
Suzuki T, Shin-I T, Fujiyama A et al (2005) Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol 174:2885–2891
Suzuki T, Shin-I T, Kohara Y, Kasahara M (2004) Transcriptome analysis of hagfish leukocytes: a framework for understanding the immune system of jawless fishes. Dev Comp Immunol 28:993–1003
Taddese B, Garnier A, Deniaud M et al (2021) Bios2cor: an R package integrating dynamic and evolutionary correlations to identify functionally important residues in proteins. Bioinformatics 37:2483–2484
Tanaka S, Baba Y (2020) B cell receptor signaling. Adv Exp Med Biol 1254:23–36
Thompson AW, Hawkins MB, Parey E et al (2021) The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat Genet 53:1373–1384
Timms JF, Carlberg K, Gu H et al (1998) Identification of major binding proteins and substrates for the SH2-containing protein tyrosine phosphatase SHP-1 in macrophages. Mol Cell Biol 18:3838–3850
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302:575–581
Tregaskes CA, Kong FK, Paramithiotis E et al (1995) Identification and analysis of the expression of CD8 alpha beta and CD8 alpha alpha isoforms in chickens reveals a major TCR-gamma delta CD8 alpha beta subset of intestinal intraepithelial lymphocytes. J Immunol 154:4485–4494
Tseng J, Lee YJ, Eisfelder BJ, Clark MR (1994) The B cell antigen receptor complex: mechanisms and implications of tyrosine kinase activation. Immunol Res 13:299–310
Tsuchiya Y, Mizuguchi K (2016) The diversity of H3 loops determines the antigen-binding tendencies of antibody CDR loops. Protein Sci 25:815–825
Tukwasibwe S, Nakimuli A, Traherne J et al (2020) Variations in killer-cell immunoglobulin-like receptor and human leukocyte antigen genes and immunity to malaria. Cell Mol Immunol 17:799–806
Uhrberg M, Parham P, Wernet P (2002) Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54:221–229
van Beek EM, Cochrane F, Barclay AN, van den Berg TK (2005) Signal regulatory proteins in the immune system. J Immunol 175:7781–7787
van den Berg TK, Yoder JA, Litman GW (2004) On the origins of adaptive immunity: innate immune receptors join the tale. Trends Immunol 25:11–16
Vernon-Wilson EF, Kee WJ, Willis AC et al (2000) CD47 is a ligand for rat macrophage membrane signal regulatory protein SIRP (OX41) and human SIRPalpha 1. Eur J Immunol 30:2130–2137
Viertlboeck BC, Göbel TW (2011) The chicken leukocyte receptor cluster. Vet Immunol Immunopathol 144:1–10
Viertlboeck BC, Schmitt R, Göbel TW (2006) The chicken immunoregulatory receptor families SIRP, TREM, and CMRF35/CD300L. Immunogenetics 58:180–190
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Vitallé J, Terrén I, Orrantia A et al (2019) CD300 receptor family in viral infections. Eur J Immunol 49:364–374
Wcisel DJ, Howard JT 3rd, Yoder JA, Dornburg A (2020) Transcriptome ortholog alignment sequence tools (TOAST) for phylogenomic dataset assembly. BMC Evol Biol 20:41
Wcisel DJ, Ota T, Litman GW, Yoder JA (2017) Spotted gar and the evolution of innate immune receptors. J Exp Zool B Mol Dev Evol 328(7):666–684
Wcisel DJ, Yoder JA (2016) The confounding complexity of innate immune receptors within and between teleost species. Fish Shellfish Immunol 53:24–34
Wei S, Zhou J-M, Chen X et al (2007) The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 59:813–821
Yamanoue Y, Miya M, Doi H, et al (2011) Multiple invasions into freshwater by pufferfishes (teleostei: tetraodontidae): a mitogenomic perspective. PLoS One 6:e17410
Yanagi Y, Yoshikai Y, Leggett K et al (1984) A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature 308:145–149
Yoder JA (2009) Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144
Yoder JA, Cannon JP, Litman RT et al (2008) Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics 60:257–265
Yoder JA, Litman GW (2000) Immune-type diversity in the absence of somatic rearrangement. Curr Top Microbiol Immunol 248:271–282
Yoder JA, Litman GW (2011) The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63:123–141
Yoder JA, Litman RT, Mueller MG et al (2004) Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A 101:15706–15711
Yoder JA, Mueller MG, Wei S et al (2001) Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci U S A 98:6771–6776
Yoder JA, Mueller MG, Nichols KM, Ristow SS, Thorgaard GH, Ota T, Litman GW (2002) Cloning novel immune-type inhibitory receptors from the rainbow trout, Oncorhynchus mykiss. Immunogenetics 54:662–670
Zhang Y, Xu K, Deng A et al (2014) An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1. Proc Natl Acad Sci U S A 111:397–402
Acknowledgements
We thank Gary Litman (University of South Florida, retired) for decades of discussions on the evolution of immunity and the role NITRs may play in Actinopterygii, Martin Flajnik (University of Maryland at Baltimore) for critical reading of the manuscript and sharing his extensive knowledge of chondrichthyan immune receptors, and Thomas Near (Yale University) for numerous discussions about the evolutionary history of ray-finned fishes and for providing the illustrations used in Fig. 4. Finally, we would like to thank Kara Carlson (North Carolina State University), April Lamb (University of North Carolina at Charlotte), Rittika Mallik (University of North Carolina at Charlotte), Cameron Nguyen (University of North Carolina at Charlotte), Drake Phelps (North Carolina State University), and Katerina Zapfe (University of North Carolina at Charlotte) for reading and providing helpful comments on the earlier drafts of this manuscript
Funding
This report was supported, in part, by grants from the National Science Foundation (IOS-1755242 to AD and IOS-1755330 to JAY) and from the Triangle Center for Evolutionary Medicine (TriCEM) to AD and JAY.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dornburg, A., Yoder, J.A. On the relationship between extant innate immune receptors and the evolutionary origins of jawed vertebrate adaptive immunity. Immunogenetics 74, 111–128 (2022). https://doi.org/10.1007/s00251-021-01232-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01232-7