Abstract
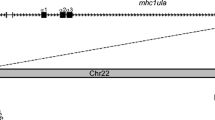
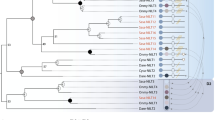
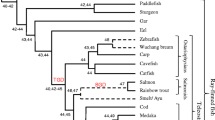
Bony fish encode multiple multi-gene families of membrane receptors that are comprised of immunoglobulin (Ig) domains and are predicted to function in innate immunity. One of these families, the diverse immunoglobulin (Ig) domain-containing protein (DICP) genes, maps to three chromosomal loci in zebrafish. Most DICPs possess one or two Ig ectodomains and include membrane-bound and secreted forms. Membrane-bound DICPs include putative inhibitory and activating receptors. Recombinant DICP Ig domains bind lipids with varying specificity, a characteristic shared with mammalian CD300 and TREM family members. Numerous DICP transcripts amplified from different lines of zebrafish did not match the zebrafish reference genome sequence suggesting polymorphic and haplotypic variation. The expression of DICPs in three different lines of zebrafish has been characterized employing PCR-based strategies. Certain DICPs exhibit restricted expression in adult tissues whereas others are expressed ubiquitously. Transcripts of a subset of DICPs can be detected during embryonic development suggesting roles in embryonic immunity or other developmental processes. Transcripts representing 11 previously uncharacterized DICP sequences were identified. The assignment of two of these sequences to an unplaced genomic scaffold resulted in the identification of an alternative DICP haplotype that is linked to a MHC class I Z lineage haplotype on zebrafish chromosome 3. The linkage of DICP and MHC class I genes also is observable in the genomes of the related grass carp (Ctenopharyngodon idellus) and common carp (Cyprinus carpio) suggesting that this is a shared character with the last common Cyprinidae ancestor.








Similar content being viewed by others
References
Abi Rached L, McDermott MF, Pontarotti P (1999) The MHC big bang. Immunol Rev 167:33–44
Aparicio S, Chapman J, Stupka E et al (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310. doi:10.1126/science.1072104
Berry R, Ng N, Saunders PM et al (2013) Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat Immunol 14:699–705. doi:10.1038/ni.2605
Borrego F (2013) The CD300 molecules: an emerging family of regulators of the immune system. Blood 121:1951–1960. doi:10.1182/blood-2012-09-435057
Broughton RE, Betancur-R R, Li C, Arratia G, Ortí G (2013) Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr 16:5. doi:10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e
Brown KH, Dobrinski KP, Lee AS et al (2012) Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci 109:529–534. doi:10.1073/pnas.1112163109
Cannon JP, O’Driscoll M, Litman GW (2012) Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics 64:39–47. doi:10.1007/s00251-011-0562-4
Clark GJ, Ju X, Tate C, Hart DN (2009) The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol 30:209–217. doi:10.1016/j.it.2009.02.003
Dirscherl H, Yoder JA (2014) Characterization of the Z lineage major histocompatability complex class I genes in zebrafish. Immunogenetics 66:185–198. doi:10.1007/s00251-013-0748-z
Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11(1):47–59. doi:10.1038/nrg2703
Frischmeyer-Guerrerio PA, Montgomery RA, Warren DS et al (2011) Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc Natl Acad Sci U S A 108:10638–10643. doi:10.1073/pnas.1019352108
Gloggnitzer J, Akimcheva S, Srinivasan A et al (2014) Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16:376–390. doi:10.1016/j.chom.2014.08.010
Haire RN, Cannon JP, O’Driscoll ML et al (2012) Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics 99:282–291. doi:10.1016/j.ygeno.2012.02.004
Hamid FM, Makeyev EV (2014) Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem Soc Trans 42:1168–1173. doi:10.1042/BST20140066
Hartmann J, Tran TV, Kaudeer J et al (2012) The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. J Biol Chem 287:31527–31539. doi:10.1074/jbc.M111.304238
Howe K, Clark MD, Torroja CF et al (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. doi:10.1038/nature12111
Jackson AN, McLure CA, Dawkins RL, Keating PJ (2007) Mannose binding lectin (MBL) copy number polymorphism in Zebrafish (D. rerio) and identification of haplotypes resistant to L. anguillarum. Immunogenetics 59:861–872. doi:10.1007/s00251-007-0251-5
Jima DD, Shah RN, Orcutt TM et al (2009) Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol Immunol 46:1505–1516. doi:10.1016/j.molimm.2008.12.021
Kalyna M, Simpson CG, Syed NH et al (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40:2454–2469. doi:10.1093/nar/gkr932
Kasahara M (1999) The chromosomal duplication model of the major histocompatibility complex. Immunol Rev 167:17–32
Kervestin S, Jacobson A (2012) NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13:700–712. doi:10.1038/nrm3454
Konantz M, Balci TB, Hartwig UF et al (2012) Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci 1266:124–137. doi:10.1111/j.1749-6632.2012.06575.x
Kortum AN, Rodriguez-Nunez I, Yang J et al (2014) Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 66:267–279. doi:10.1007/s00251-014-0759-4
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–5. doi:10.1093/nar/gkr931
Loucks E, Carvan MJ 3rd (2004) Strain-dependent effects of developmental ethanol exposure in zebrafish. Neurotoxicol Teratol 26:745–755. doi:10.1016/j.ntt.2004.06.017
Maisey K, Imarai M (2011) Diversity of teleost leukocyte molecules: role of alternative splicing. Fish Shellfish Immunol 31:663–672. doi:10.1016/j.fsi.2010.08.001
Meeker ND, Hutchinson SA, Ho L, Trede NS (2007) Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 43:610–614. doi:10.2144/000112619
Moody AM, Chui D, Reche PA, Priatel JJ, Marth JD, Reinherz EL (2001) Developmentally regulated glycosylation of the CD8alphabeta coreceptor stalk modulates ligand binding. Cell 107:501–512. doi:10.1016/S0092-8674(01)00577-3
Novoa B, Figueras A (2012) Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol 946:253–275. doi:10.1007/978-1-4614-0106-3_15
Ohashi K, Takizawa F, Tokumaru N et al (2010) A molecule in teleost fish, related with human MHC-encoded G6F, has a cytoplasmic tail with ITAM and marks the surface of thrombocytes and in some fishes also of erythrocytes. Immunogenetics 62:543–559. doi:10.1007/s00251-010-0460-1
Olsson LM, Holmdahl R (2012) Copy number variation in autoimmunity—importance hidden in complexity? Eur J Immunol 42:1969–1976. doi:10.1002/eji.201242601
Patowary A, Purkanti R, Singh M et al (2013) A sequence-based variation map of zebrafish. Zebrafish 10:15–20. doi:10.1089/zeb.2012.0848
Pelak K, Need AC, Fellay J et al (2011) Copy number variation of KIR genes influences HIV-1 control. PLoS Biol 9:e1001208. doi:10.1371/journal.pbio.1001208
Pelham CJ, Agrawal DK (2014) Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert Rev Clin Immunol 10:243–256. doi:10.1586/1744666X.2014.866519
Pimentel H, Parra M, Gee S et al (2014) A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res 42:4031–4042. doi:10.1093/nar/gkt1388
Renshaw SA, Trede NS (2012) A model 450 million years in the making: zebrafish and vertebrate immunity. Dis Model Mech 5:38–47. doi:10.1242/dmm.007138
Rodriguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA (2014) Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: deciphering the differences. Dev Comp Immunol 46:24–34. doi:10.1016/j.dci.2014.02.004
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakai M, Kono T, Savan R (2005) Identification of expressed genes in carp (Cyprinus carpio) head kidney cells after in vitro treatment with immunostimulants. Dev Biol (Basel) 121:45–51
Sipes NS, Padilla S, Knudsen TB (2011) Zebrafish: as an integrative model for twenty-first century toxicity testing. Birth Defects Res C Embryo Today 93:256–267. doi:10.1002/bdrc.20214
Stafford JL, Bengten E, Du Pasquier L et al (2006) A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 58:758–773. doi:10.1007/s00251-006-0134-1
Star B, Nederbragt AJ, Jentoft S et al (2011) The genome sequence of Atlantic cod reveals a unique immune system. Nature 477:207–210. doi:10.1038/nature10342
Stet RJ, Hermsen T, Westphal AH et al (2005) Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics 57:77–89. doi:10.1007/s00251-005-0771-9
Sullivan C, Kim CH (2008) Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol 25:341–350. doi:10.1016/j.fsi.2008.05.005
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Traherne JA, Martin M, Ward R et al (2010) Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet 19:737–751. doi:10.1093/hmg/ddp538
Traver D (2004) Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol 76:127–149
Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI (2003) Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 4:1238–1246. doi:10.1038/ni1007
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115, doi:10.1093/nar/gks596
Veldman MB, Lin S (2008) Zebrafish as a developmental model organism for pediatric research. Pediatr Res 64:470–476. doi:10.1203/PDR.0b013e318186e609
Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S (2008) Functions of natural killer cells. Nat Immunol 9(5):503–10. doi:10.1038/ni1582
Wang Y, Lu Y, Zhang Y et al (2015) The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet 47(6):625–31. doi:10.1038/ng.3280
Wei S, Zhou JM, Chen X et al (2007) The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 59(10):813–21. doi:10.1007/s00251-007-0250-6
Westerfield M (2007) The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). Univ. of Oregon Press, Eugene
Wittamer V, Bertrand JY, Gutschow PW, Traver D (2011) Characterization of the mononuclear phagocyte system in zebrafish. Blood 117:7126–7135. doi:10.1182/blood-2010-11-321448
Wong JJ, Ritchie W, Ebner OA et al (2013) Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154:583–595. doi:10.1016/j.cell.2013.06.052
Xu C, Call ME, Wucherpfennig KW (2006) A membrane-proximal tetracysteine motif contributes to assembly of CD3deltaepsilon and CD3gammaepsilon dimers with the T cell receptor. J Biol Chem 281:36977–36984. doi:10.1074/jbc.M607164200
Xu P, Zhang X, Wang X et al (2014) Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet 46(11):1212–9. doi:10.1038/ng.3098
Yang L, Zou M, Fu B, He S (2013) Genome-wide identification, characterization, and expression analysis of lineage-specific genes within zebrafish. BMC Genomics 14:65–2164-14-65. doi:10.1186/1471-2164-14-65
Yoder JA, Turner PM, Wright PD et al (2010) Developmental and tissue-specific expression of NITRs. Immunogenetics 62:117–122. doi:10.1007/s00251-009-0416-5
Yoder JA (2009) Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144. doi:10.1016/j.dci.2008.09.004
Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW (2008) Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics 60:257–265. doi:10.1007/s00251-008-0285-3
Yoder JA, Litman RT, Mueller MG et al (2004) Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A 101:15706–15711, 0405242101
Yoder JA, Mueller MG, Wei S et al (2001) Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci U S A 98:6771–6776. doi:10.1073/pnas.121101598
Acknowledgment
We thank Hayley Dirscherl (NC State University) for helpful discussions and zebrafish genomic DNA samples, John Whitesides and Patti McDermott (Duke Human Vaccine Institute, Duke University School of Medicine) for assistance with cell sorting, Diana Skapura (All Children’s Hospital) for assistance with DNA sequencing, and Barbara Pryor for editorial assistance. The authors are supported by the National Institutes of Health (R01 AI057559 to G.W.L. and J.A.Y. and R01 AI23337 to G.W.L.) and the National Evolutionary Synthesis Center (NESCent), NSF #EF‐0905606 (D.J.W.).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
Online Resource 1. Table S1. Complete DICP amplicon sequence data including cDNA source, DNA polymerase, nucleotide and amino acid sequence, and GenBank accession numbers. (XLSX 45 kb)
ESM 2
Online Resource 2. Table S2. Percent (nucleotide) identity between DICP amplicons generated with a high-fidelity DNA polymerase and previously described DICP sequences. (XLSX 17 kb)
ESM 3
Online Resource 3. DICP sequence variation; DICP exon-intron architecture Figures S1–S14; Tables S3–S4. (PDF 2796 kb)
Rights and permissions
About this article
Cite this article
Rodriguez-Nunez, I., Wcisel, D.J., Litman, R.T. et al. The identification of additional zebrafish DICP genes reveals haplotype variation and linkage to MHC class I genes. Immunogenetics 68, 295–312 (2016). https://doi.org/10.1007/s00251-016-0901-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-016-0901-6