Abstract
Background
To analyze the expression of miRNA (microRNA) in peripheral blood mononuclear cells in patients with Psoriasis vulgaris with different TCM syndromes by miRNA chip. It further revealed the micromaterial basis of different syndrome types of psoriasis at the miRNA level.
Methods
Peripheral blood monocytes were collected and prepared from 30 patients with Psoriasis vulgaris (including 9 patients of blood heat syndrome, 8 patients of blood stasis syndrome, and 13 patients of blood dry syndrome) and 9 healthy controls. The miRNA expression profile of peripheral blood monocytes was detected by Agilent Hum miRNA chip.
Results
Compared to the healthy control group, 156 upregulated and 242 downregulated miRNAs were detected in all psoriasis patients. Compared to the healthy control group, 40 miRNAs were upregulated and 44 were downregulated in the blood heat syndrome group. Furthermore, there were 49 upregulated miRNAs and 44 downregulated miRNAs in the dry syndrome group as compared to the healthy control group. Also, 67 miRNAs were upregulated and 154 miRNAs were downregulated in the blood stasis syndrome group as compared to the healthy control group.
Conclusions
There are common different miRNAs and pathways, as well as specific miRNAs between the psoriasis and the healthy control groups.
Trial registration ChiCTR-TRC-14005185 registered on August 8, 2014.
Similar content being viewed by others
Background
Psoriasis is a chronic and recurrent skin inflammatory disease. In 2016, the World Health Organization (WHO) reported that the global prevalence of psoriasis ranged from 0.09 to 11.43% [1], and the incidence of psoriasis in China has increased from 0.12 to 1.49% in the past 25 years. Psoriasis has become a common disease endangering public health. Currently, there are defects in the treatment of psoriasis, such as a short remission period; also, the disease is easy to relapse or tolerate. Moreover, most drugs have considerable side effects. Traditional Chinese Medicine (TCM) treatment of psoriasis has the advantages of a long remission period, stable condition, not easy to rebound, and less toxicity and side effects. In recent years, it has attracted the attention of the medical community worldwide. Traditional Chinese medicine (TCM) has a unique theoretical system in the treatment of psoriasis and has accumulated abundant clinical experience for a prolonged period. TCM syndrome is the core of clinical TCM and vital for the modernization of TCM. Our research group analyzed the distribution of Psoriasis vulgaris-related literature from 1979 to 2010, and found that among all the syndromes of the disease, the top three were blood heat syndrome (32.86%), blood dry syndrome (23.56%), and blood stasis syndrome (19.43%), accounting for a total of 75.85% [2]. Moreover, blood stasis syndrome is related to a stable period, while the regression period is related to the blood dry syndrome [3].
Precision medicine is based on individualized medicine; it uses genomics, proteomics, and data analysis to achieve personalized and precise prevention and treatment of the diseases [4]. The connotation coincides with the doctrine “treatment based on syndrome differentiation” in traditional Chinese medicine. It can summarize the classification of TCM syndrome differentiation at the microscopic and molecular level based on the combination of genomics and proteomics to achieve accurate diagnosis and treatment as well as improve the therapeutic effect.
No correlation has been established between TCM classification of psoriasis and miRNA omics. In this study, we analyzed the differences of miRNA expression in common TCM types of psoriasis (blood heat syndrome, blood dry syndrome, and blood stasis syndrome) by miRNA microarray method and revealed the microscopic variations of the different syndrome types of psoriasis at the miRNA level.
Materials and methods
Recruitment and characteristics of patients
All patients were from the Dermatology Clinic and Ward of Guangdong Hospital of Traditional Chinese Medicine. This study was approved by the ethics committee of our hospital, and all patients and healthy individuals signed the informed consent. Psoriasis vulgaris group consisted of 30 patients, including 9 patients of blood heat syndrome, 8 patients of blood stasis syndrome, and 13 patients of the dry syndrome. The age of the patients was from 19 to 60 years, the duration of the disease was 1–10 years, the diagnostic standard was consistent with Psoriasis vulgaris [5], and the diagnosis of TCM syndrome was consistent with the diagnosis of bai bi syndrome [6]. The healthy control group comprised of 9 healthy patients as assessed in an outpatient physical examination and matched with the psoriasis group in gender and age. All the human studies were in accordance with the Helsinki declaration and Chinese relevant clinical trial research standards and regulations. All subjects included in this study were introduced to the purpose and treatment of this study by the researcher before specimen collection.
Exclusion criteria
Pregnant or lactating women; patients administered steroids and/or retinoids or biological agents in the last six months; patients who used steroids or retinoids within 1 month; patients with primary diseases such as hyperthyroidism, cardiovascular, cerebrovascular, liver, kidney and hematopoietic system, as well as those on long-term medication; a variety of Chinese medicine syndrome type inclusion of patients.
Instruments and reagents
TRI Reagent (catalogue no. T9424; Sigma, Germany), miRNeasy Micro Kit (cat alogue no. T9424 217084; Qiagen, Germany), RNase-Free DNase Set (cat alogue no. 79254; Qiagen), Tiangen miRcute miRNA cDNA First-Strand Synthesis Kit (cat alogue no. KR201; Tiangen, Beijing, China), SYBR Green PCR kit (cat alogue no. Fp411-02; Tiangen, Beijing, China), Agilent Bioanalyzer 2100 analyzer (Agilent Technologies, Santa Clara, CA, USA); Scanner G2505C (Agilent Technologies), feature extraction (version 10.7.1.1, Agilent Technologies), data processing software Genespring (version 13.1, Agilent Technologies), and Affymetrix Agilent Human miRNA, Release 21.0 (8 × 60K, Design ID: 070156) chip were utilized for this study.
Sample collection and processing
A volume of 10 mL elbow vein blood was collected in an anticoagulant tube containing heparin, and peripheral blood monocytes (PBMCs) were separated according to the manufacturer’s instructions and stored at − 80 °C.
Extraction and quality control of RNA
Total RNA was extracted using TRI reagent according to the manufacturer’s protocol and purified using the miRNeasy Micro Kit and RNase-Free DNase Set. During each RNA extraction experiment, a control total RNA was utilized as positive quality control and RNase-free water as negative quality control. After quality control (QA) evaluation, the integrity of the purified total RNA was detected by Agilent Bioanalyzer 2100 (Agilent technologies).
Agilent miRNA microarray experiment
The chip used in this study was Agilent Human miRNA, Release 21.0 (8 × 60K, Design ID: 070156), and a total of 39 samples were on-chip and analyzed. Total RNA was quantified by NanoDrop ND-2000 (Thermo Scientific, MA, USA) and detected by Agilent Bioanalyzer 2100. After passing the RNA quality inspection, the samples were labeled, hybridized, and eluted according to the standard chip procedure. First, total RNA was dephosphorylated and denatured, followed by labeling with cyanine-3-CTP (Cy3). The labeled RNA was purified and hybridized with the chip. After elution, the original image was scanned by Agilent Scanner G2505C.
Feature Extraction software was used to extract the original data from the original image, and Genespring software was utilized for standardization and subsequent processing. The standardized data were filtered, and at least one set of 100% probes labeled “Detected” from each set of samples used for comparison was stored for subsequent analysis. P-values and multiple change values of T-test were used to screen differential miRNAs, and the screening criteria were upregulated or downregulated multiple change value ≥ 2.0 and P ≤ 0.05. Then, three databases (TargetScan, microRNA.org, PITA) were used to jointly predict the target genes of different miRNAs, followed by GO and KEGG enrichment analysis of the target genes to determine the biological functions or pathways mainly affected by different miRNAs. Finally, unsupervised hierarchical clustering was carried out for different miRNAs, and the expression patterns of different miRNAs in different samples were displayed in the form of a heatmap.
Validation by quantitative real-time polymerase chain reaction (qRT-PCR)
The isolated RNA from the PBMC samples from different groups was reverse transcribed directly into cDNA using a SuperScript IV Reverse Transcriptase (Thermo Fisher, Dalian, China) according to the manufacturer’s instructions. PCR amplification conditions were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 94 °C for 20 s and annealing at 60 °C for 34 s. The expression of microRNAs was examined using qRT-PCR by a SYBR Green PCR Kit.
Statistical analysis
Statistical analyses were performed using SPSS software 19.0 and represented using GraphPad Prism 5.0 (La Jolla, CA, USA). The results are expressed as means ± standard errors of the means. Student’s t-test was used for the statistical analysis. P-values < 0.05 were considered significant.
Results
Quality control of RNA chip data is satisfactory
Combined with the clinical sample information, we selected 39 samples (9 samples of hematic psoriasis, 13 samples of dry psoriasis, 8 samples of blood stasis psoriasis, and 9 samples of the control group) to perform Agilent miRNA microarray analysis. The quality control of the original result data was satisfactory (Fig. 1).
Analysis of miRNA microarray results of blood heat syndrome and control groups
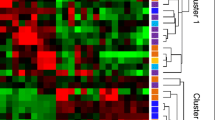
Next, we conducted a preliminary analysis of miRNA microarray results from the blood heat syndrome and the control groups, and miRNAs with fold-change > 2 and P-value < 0.05 were screened out. Among these differential miRNAs, 40 were upregulated and 44 were downregulated. The Volcano Map was used to reflect the differences in the expression of miRNAs between the blood heat syndrome and the control groups (Fig. 2). The heatmap displayed the results of unsupervised hierarchical clustering of miRNAs differences between both groups (Fig. 2). In addition, Venn analysis of the differentially expressed genes compared the miRNA microarray data from both groups (Fig. 3).
According to the results of the miRNA microarray, the first three upregulated miRNAs were miR-32-3p, miR-144-3p, and -miR-574-3p, and the first two downregulated miRNAs were miR-638 and miR-572. The verification results of qRT-PCR were consistent with those of miRNA microarray (Fig. 4).
Analysis of miRNA microarray results of psoriasis blood dry syndrome and control groups
Herein, we conducted a preliminary analysis of miRNA microarray results in the blood dry syndrome and the control groups and miRNAs with fold-change > 2 and P-value < 0.05 screened out. Among these differential miRNAs, 49 were upregulated and 44 were downregulated. The Volcano Map reflected the differences in the expression of miRNAs between the blood dry syndrome and the control groups (Fig. 2), while the Heatmap showed the results of unsupervised hierarchical clustering of miRNA differences between both control group (Fig. 2). We also performed a Venn analysis of the differentially expressed genes to compare the miRNA microarray data in the blood dry syndrome and control groups (Fig. 3).
According to the results of miRNA microarrays, the first four upregulated miRNAs were miR-144-3p,miR-32-3p, miR-574-3p, and miR-466, and the first two down-regulated miRNAs were miR-638 and miR-572, and the verification results of qRT-PCR were consistent with the results of miRNA microarray (Fig. 4).
Analysis of miRNA microarray results of psoriasis blood stasis syndrome and control groups
A preliminary analysis of miRNA microarray results was conducted in the blood stasis syndrome and the control groups, and miRNAs with fold-change > 2 and P-value < 0.05 were screened out. Among these differential miRNAs, 67 were upregulated and 154 were downregulated. The Volcano Map was used to reflect the differences in the expression of miRNAs between the blood stasis syndrome and the control groups (Fig. 2), and Heatmap showed the results of unsupervised hierarchical clustering of miRNAs differences between both groups (Fig. 2). Next, a Venn analysis of the differentially expressed genes was used to compare the miRNA microarray data from both group (Fig. 3).
According to the results of miRNA microarray, the first two upregulated miRNAs were miR-32-3p and miR-483-3p, and the first three downregulated miRNAs were miR-138-2-3p, miR-378b, and miR-142-3p; also, the verification results of qRT-PCR were consistent with those of miRNA microarray (Fig. 4).
miRNA-enrichment pathways and of the three psoriasis syndromes were analyzed from the control group
After the target gene prediction of the different miRNAs in the control group, we used three databases (TargetScan, microRNA.org, and PITA) to enrich the pathways for the three psoriasis syndromes. Venn analysis was performed on the pathways of the three syndromes (Fig. 3).
As seen from Table 1, the unique differential pathway in the psoriatic blood heat syndrome group was pancreatic secretion, while that in the psoriatic blood dry syndrome group included Shigellosis, bacterial invasion of epithelial cells, endocrine, and other factor-regulated calcium reabsorption and Th17 cell differentiation. The unique differential pathway in the psoriatic blood stasis syndrome group included human T-cell leukemia virus 1 infection, calcium signaling pathway, and ECM-receptor interaction.
Furthermore, the DIANA TOOLS online tool and its three databases (TarBase v7.0, microt-cds (v 5.0) and TargetScan) were used to correlate the differentially expressed miRNAs and genes (Fig. 5).
Correlation analysis of differentially expressed miRNAs and genes between three syndromes of psoriasis and healthy control groups. a Correlation analysis of differentially expressed miRNAs and genes between blood-heat syndrome and healthy control groups. b Correlation analysis of differentially expressed miRNAs and genes between blood-stasis syndrome and healthy control groups. c Correlation analysis of differentially expressed miRNAs and genes between blood-dryness syndrome and healthy control groups
Discussion
In this study, miRNA microarray was used to analyze the three most common TCM types of psoriasis at the level of miRNA expression. First, a preliminary analysis of miRNA microarray results was conducted and miRNAs with fold-change > 2 and P-value < 0.05 were screened out. Among these differential miRNAs, 40 were upregulated and 44 were downregulated in patients with blood heat, 49 were upregulated and 44 were downregulated in patients with blood dry, and 67 were upregulated and 154 were downregulated in patients with blood stasis as compared to the normal group. The comparison of the miRNA microarray data of the three psoriasis types of blood heat, blood stasis, and blood dry to that of the control group found that the three types of psoriasis had unique but different miRNAs as well as common miRNAs.
In the current analysis, compared to the control group, we found that the three syndromes of psoriasis had a common upregulated different miRNA: mir-32-3p. Reportedly, this miRNA is related to malignant pleural mesothelioma, acute ischemic stroke, chronic rhinosinusitis, and is also a potential biomarker [7,8,9]. The upregulation of miR-144-3p in both hematopoiesis and dry inhibits cell proliferation by negative regulation of the expression of Tie2 in pulmonary microvascular endothelial cells (PMVECs), and the overexpression of miR-144-3p can be used in the treatment of hepatopulmonary syndrome (HPS) [10]. In addition, miR-144-3p is significantly increased in the bone marrow and peripheral blood of patients with acute myeloid leukemia. miR-144-3p promotes the proliferation of HL-60 cells by inhibiting NRF2. Therefore, miR-144-3p can be considered as a non-invasive biological marker of leukemia [11]. The expression of miR-574-3p is upregulated in the syndrome of blood heat and dry, and the automatic regulation feedback loop of iASPP/p63 requires miR-574-3p to regulate homeostasis of the tunable and stratified epithelium [12]. In addition, miR-574-3p is also involved in the adipogenesis of the sebaceous gland [13]. The upregulation of miR-466 in dry syndrome is related to host immunity in the skin [14] and is involved in skin and mucosal HPV infection [15]. The upregulation of miR-483-3p in blood stasis syndrome is significantly increased in the plasma of pancreatic cancer patients [16], which suggests poor prognosis in pancreatic duct adenocarcinoma patients [17]. The miR-638 and miR-572 are significantly downregulated in basal cell carcinoma [18], suggesting their involvement in the pathogenesis of the disease. The downregulated miR-378b in blood stasis syndrome promotes keratinocyte differentiation by acting on NKX3.1 [19]. In addition, the downregulated miR-142-3p in blood stasis syndrome is a RANKL-dependent inducer of osteoclast death [20].
Next, we predicted the different miRNA target genes between the three syndromes and the control group and carried out KEGG enrichment. After Venn analysis on the pathway, we found that the three syndromes had a common as well as a specific pathway. The common different pathways of the three syndromes, signaling pathways regulating the pluripotency of stem cells, needed the activation of MAPK and PI3K signaling pathways that are closely related to the occurrence of psoriasis [21]. A PI3K-Akt signaling pathway is associated with the occurrence and development of psoriasis. A previous study reported that PI3K signaling regulates the proliferation of keratinocytes by activating Akt and other targets and inducing the downregulation of FOXO [22]. In addition, the PI3K-Akt-mTOR pathway induces keratinocyte proliferation and inflammatory cytokine secretion by inhibiting autophagy in keratinocytes. The inhibition of the PI3K-Akt-mTOR pathway alleviates the psoriasis symptoms [23,24,25,26,27,28]. The common different pathways of the three syndromes include the MAPK signaling pathway. The MAPK signaling pathway is a serine/threonine protein kinase, which mainly includes extracellular signal-regulated protein kinases (ERK), C-jun N-terminal kinases (JNK), and p38 MAPK. Some studies have shown that excessive activation of p38 MAPK leads to decreased function of the skin barrier in psoriasis as well as skin inflammation [29,30,31,32].
The common pathway of the three syndromes is axon guidance, which is involved in the occurrence and development of psoriasis. Reportedly, the axonal guide molecules, CD100 and Plexin-B2, are involved in the inflammatory process of psoriasis by activating NF-κB and NLRP3 inflammasome in keratinocytes [33].
The specific pathway of dry psoriasis includes Th17 cell differentiation. Genetic studies have reinforced that Th17 cells play a central role in psoriasis [34]. These cells are responsible for the production of the cytokine IL-17A. TGF- β with IL -6 or IL -21 causes initial CD4+ T cells to differentiate into the Th17 subgroup, and IL -23, which is the growth-stabilizing factor that affects the normal secretion process of IL-17. A large number of Th17 cells were infiltrated into the lesions of psoriasis. Lewis et al. [35] isolated and counted Th17 cells in the plaque tissues of patients, and found the proportion of CD4+ Th cells was 49–93%. Th17 cells exert an effect by secreting IL-17A, IL-17F, IL-26, and TNF in psoriasis, forming a feedforward inflammatory network with KCs, activating STAT-1 and NF-κB signaling pathway, and inducing keratinocytes (KCs) to secrete several pro-inflammatory factors. IL-17 and TNF can jointly stimulate the high expression of KCs CCL20 [36] to collect the activated CD11c + macrophage-derived dendritic cells (mDCs) and adjacent CCR6 + T17 cells and induce the differentiation of T cells in skin drainage lymph nodes towards Th17 and Tc17.
The human T-cell leukemia 1 infection pathway is abnormal in blood stasis psoriasis. A previous study showed that ustekinumab could successfully treat psoriasis with human -cell leukemia/type 1 lympho virus infection [37]. Further analysis found that osteoclast differentiation, a unique pathway, was abnormal in blood stasis psoriasis. Raimondo et al. demonstrated that skin inflammation in psoriasis promoted the differentiation of human monocytes into active osteoclasts and bone injury [38].
Conclusions
In this study, we connected three common TCM syndromes of psoriasis with miRNAs and provided a reference for understanding the TCM syndromes of psoriasis. In the future study, we will explore the mechanism of the differentially expressed miRNAs from different TCM syndromes.
Availability of data and materials
The datasets used and/or analyzed during the current study would be available from the corresponding author on reasonable request.
Abbreviations
- TCM:
-
Traditional Chinese Medicine
- PBMCs:
-
Peripheral blood mononuclear cells
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- miRNA:
-
microRNA
- PMVECs:
-
Pulmonary microvascular endothelial cells
- HPS:
-
Hepatopulmonary syndrome
- ERK:
-
Extracellular signal-regulated protein kinases
- JNK:
-
C-jun N-terminal kinases
- KCs:
-
Keratinocytes
- mDCs:
-
Macrophage-derived dendritic cells
References
World Health Organization. Global report on psoriasis. Geneva: WHO; 2016. p. 7–9.
Lu CJ, Zeng Z, Xie XL, Ning J. Distribution of Chinese Medical Syndrome in ordinary psoriasis: literature from 1979 to 2010. J Tradit Chin Med. 2012;53(11):959–61 (Article in Chinese).
Yan YH, Lu CJ, Yao DN, Du H, Li HL. Study on the correlation between the basic Syndrome of traditional Chinese Medicine and the stage and condition of Psoriasis vulgaris. J Guangzhou Univ Tradit Chin Med. 2012;04:22–6 (Article in Chinese).
Li XH, Jia LL, Liu XX. Preliminary discussion of diagnosis and treatment mode of future medicine from treating based on syndrome differentiation and precision medicine. Chin J Tradit Chin Med. 2018;33(11):4789–92 (Article in Chinese).
Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386(9997):983–94.
Lu CJ, Yu JJ, Deng JW. Disease-syndrome combination clinical study of psoriasis: present status, advantages, and prospects. Chin J Integrat Med. 2012;18(3):166–71.
Xuan L, Luan G, Wang Y, Lan F, Zhang X, Hao Y, et al. MicroRNAs regulating mucin type O-glycan biosynthesis and transforming growth factor beta signaling pathways in nasal mucosa of patients with chronic rhinosinusitis with nasal polyps in Northern China. Int Forum Allergy Rhinol. 2019;9(1):106–13.
Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol. 2015;35(3):433–47.
Bononi I, Comar M, Puozzo A, Stendardo M, Boschetto P, Orecchia S, et al. Circulating microRNAs found dysregulated in ex-exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers. Oncotarget. 2016;7(50):82700–11.
Yang C, Lv K, Chen B, Yang Y, Ai X, Yu H, et al. miR144-3p inhibits PMVECs excessive proliferation in angiogenesis of hepatopulmonary syndrome via Tie2. Exp Cell Res. 2018;365(1):24–32.
Sun X, Liu D, Xue Y, Hu X. Enforced miR-144-3p expression as a non-invasive biomarker for the acute myeloid leukemia patients mainly by targeting NRF2. Clin Lab. 2017;63(4):679–87.
Chikh A, Matin RN, Senatore V, Hufbauer M, Lavery D, Raimondi C, et al. iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J. 2011;30(20):4261–73.
Schneider MR, Samborski A, Bauersachs S, Zouboulis CC. Differentially regulated microRNAs during human sebaceous lipogenesis. J Dermatol Sci. 2013;70(2):88–93.
Wu J, Gong H, Bai Y, Zhang W. Analyzing the miRNA-Gene Networks to Mine the Important miRNAs under Skin of Human and Mouse. Biomed Res Int. 2016;2016:5469371.
Gu W, An J, Ye P, Zhao KN, Antonsson A. Prediction of conserved microRNAs from skin and mucosal human papillomaviruses. Arch Virol. 2011;156(7):1161–71.
Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46(2):539–47.
Wang C, Sun Y, Wu H, Yu S, Zhang L, Meng Y, et al. Elevated miR-483-3p expression is an early event and indicates poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 2015;36(12):9447–56.
Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167(4):847–55.
Wang XL, Zhang T, Wang J, Zhang DB, Zhao F, Lin XW, et al. MiR-378b promotes differentiation of keratinocytes through NKX3.1. PLoS ONE. 2015;10(8):e0136049.
Fordham JB, Guilfoyle K, Naqvi AR, Nares S. MiR-142-3p is a RANKL-dependent inducer of cell death in osteoclasts. Sci Rep. 2016;6:24980.
Harding A, Cortez-Toledo E, Magner NL, Beegle JR, Coleal-Bergum DP, Hao D, et al. Highly efficient differentiation of endothelial cells from pluripotent stem cells requires the MAPK and the PI3K pathways. Stem Cells. 2017;35(4):909–19.
Zhang M, Zhang X. The role of PI3K/AKT/FOXO signaling in psoriasis. Arch Dermatol Res. 2019;311(2):83–91.
Lu Y, Wang AL, Zhang JW, Li L, Wei JA, Li L, et al. PSORI-CM02 ameliorates psoriasis in vivo and in vitro by inducing autophagy via inhibition of the PI3K/Akt/mTOR pathway. Phytomedicine. 2019;64:153054.
Said A, Bock S, Lajqi T, Muller G, Weindl G. Chloroquine promotes IL-17 production by CD4 + T cells via p38-dependent IL-23 release by monocyte-derived Langerhans-like cells. J Immunol. 2014;193(12):6135–43.
Wang RC, Levine B. Calcipotriol induces autophagy in HeLa cells and keratinocytes. J Invest Dermatol. 2011;131(4):990–3.
Rajawat Y, Hilioti Z, Bossis I. Autophagy: a target for retinoic acids. Autophagy. 2010;6(8):1224–6.
Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9(6):374–82.
Yang Y, Wang H, Wang S, Xu M, Liu M, Liao M, et al. GSK3beta signaling is involved in ultraviolet B-induced activation of autophagy in epidermal cells. Int J Oncol. 2012;41(5):1782–8.
Eckert RL, Efimova T, Dashti SR, Balasubramanian S, Deucher A, Crish JF, et al. Keratinocyte survival, differentiation, and death: many roads lead to mitogen-activated protein kinase. J Investig Dermatol Symp Proc. 2002;7(1):36–40.
Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68(2):320–44.
Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15(1):11–8.
Kanemaru K, Nakamura Y, Totoki K, Fukuyama T, Shoji M, Kaneko H, et al. Phospholipase Cdelta1 regulates p38 MAPK activity and skin barrier integrity. Cell Death Differ. 2017;24(6):1079–90.
Zhang C, Xiao C, Dang E, Cao J, Zhu Z, Fu M, et al. CD100-Plexin-B2 promotes the inflammation in psoriasis by activating NF-kappaB and the inflammasome in keratinocytes. J Invest Dermatol. 2018;138(2):375–83.
Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol. 2018;54(1):102–13.
Lewis BJ, Rajpara S, Haggart AM, Wilson HM, Barker RN, Ormerod AD. Predominance of activated, clonally expanded T helper type 17 cells within the CD4+ T cell population in psoriatic lesions. Clin Exp Immunol. 2013;173(1):38–46.
Zhao L, Xia J, Wang X, Xu F. Transcriptional regulation of CCL20 expression. Microbes Infect. 2014;16(10):864–70.
Kazama M, Umezawa Y, Itoh M, Nobeyama Y, Ito T, Kikuchi S, et al. Successful treatment of ustekinumab in a psoriasis patient with human T-cell leukemia/lymphotropic virus type 1 infection. J Dermatol. 2017;44(11):1334–5.
Raimondo A, Lembo S, Di Caprio R, Donnarumma G, Monfrecola G, Balato N, et al. Psoriatic cutaneous inflammation promotes human monocyte differentiation into active osteoclasts, facilitating bone damage. Eur J Immunol. 2017;47(6):1062–74.
Acknowledgements
Not applicable.
Funding
This research was financially supported by the Guangdong Province Science and Technology Planning Project (2017A050506041, 2017B030314166); the Guangdong Provincial Department of Education Project (2018KQNCX046); the Guangzhou Science and Technology Project (201807010051) and the Guangdong Provincial Hospital of Chinese Medicine Special Fund (YN2018HK01, YN2018ZD08, YN2018RBA02, YN2016XP02, YN2019QJ04, YN2019QJ08).
Author information
Authors and Affiliations
Contributions
All authors have participated extensively in the study and had proofread the final manuscript. CJL, LH and HJG conceived and designed the research. YL and YQ conducted the experiments. YHY, DNY, HD, JWD, and SYY collected patients. QBC helped sample preservation and transportation. YL and YQ analyzed the data. YL wrote the manuscript. CJL, LH and HJG approved and reviewed the final manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of Guangdong Provincial Hospital of Chinese Medicine Clinical Research Ethics Committee (Ethical approval No. B2012-53-03) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all eligible participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lu, Y., Qi, Y., Yan, Y. et al. Analysis of microRNA expression in peripheral blood monocytes of three Traditional Chinese Medicine (TCM) syndrome types in psoriasis patients. Chin Med 15, 39 (2020). https://doi.org/10.1186/s13020-020-00308-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-020-00308-y