Abstract
Background
Farnesol is an acyclic sesquiterpene alcohol present in the essential oils of various plants in nature. It has been reported to be valuable in medical applications, such as alleviation of allergic asthma, gliosis, and edema as well as anti-cancerous and anti-inflammatory effects. Coenzyme Q10 (CoQ10), an essential cofactor in the aerobic respiratory electron transport chain, has attracted growing interest owing to its clinical benefits and important applications in the pharmaceutical, food, and health industries. In this work, co-production of (E,E)-farnesol (FOH) and CoQ10 was achieved by combining 3 different exogenous terpenes or sesquiterpene synthase with the RNA interference of psy (responsible for phytoene synthesis in Rhodobacter sphaeroides GY-2).
Results
FOH production was significantly increased by overexpressing exogenous terpene synthase (TPS), phosphatidylglycerophosphatase B (PgpB), and sesquiterpene synthase (ATPS), as well as RNAi-mediated silencing of psy coding phytoene synthase (PSY) in R. sphaeroides strains. Rs-TPS, Rs-ATPS, and Rs-PgpB respectively produced 68.2%, 43.4%, and 21.9% higher FOH titers than that of the control strain. Interestingly, the CoQ10 production of these 3 recombinant R. sphaeroides strains was exactly opposite to that of FOH. However, CoQ10 production was almost unaffected in R. sphaeroides strains modified by psy RNA interference. The highest FOH production of 40.45 mg/L, which was twice as high as that of the control, was obtained from the TPS-PSYi strain, where the exogenous TPS was combined with the weakening of the phytoene synthesis pathway via psy RNA interference. CoQ10 production in TPS-PSYi, ATPS-PSYi, and PgpB-PSYi was decreased and lower than that of the control strain.
Conclusions
The original flux that contributed to phytoene synthesis was effectively redirected to provide precursors toward FOH or CoQ10 synthesis via psy RNA interference, which led to weakened carotenoid synthesis. The improved flux that was originally involved in CoQ10 production and phytoene synthesis was redirected toward FOH synthesis via metabolic modification. This is the first reported instance of FOH and CoQ10 co-production in R. sphaeroides using a metabolic engineering strategy.
Similar content being viewed by others
Background
Sesquiterpenes are C15-terpenoids composed of 3 isoprene units with numerous important medical and industrial applications [1]. (E,E)-Farnesol (FOH; C15H26O) is a colorless acyclic sesquiterpene alcohol found in many essential plant oils such as lemon grass, neroli, cyclamen, tuberose, rose, and musk [2]. FOH is known to play an important role in signal transduction [3], cell proliferation, quorum-sensing [4, 5], and apoptosis induction [6, 7]. Recently, FOH was reported to be valuable in medical application because it exhibited anti-cancerous and anti-inflammatory effects, and was also found to alleviate allergic asthma, gliosis, and edema [8, 9]. However, FOH extraction from plants is non-feasible for large scale production as only trace amounts can be found in plant essential oils. Furthermore, the plant resources for FOH production are limited to slow growth, varying composition, season, climate, and scaling-up facilitation. In order to circumvent these limitations, FOH production in prokaryotic and eukaryotic microorganisms using metabolic engineering has been exploited [10,11,12,13], along with culture optimization for enhanced FOH production using Saccharomyces cerevisiae and Candida albicans [14, 15]. Figure 1 illustrates that FOH can be obtained from the depyrophosphorylation of farnesyl pyrophosphate (FPP) catalyzed by phosphatases or sesquiterpene synthases [11]. Unfortunately, only a few studies discuss FOH production by metabolically engineering microbial hosts, even though novel medical and industrial advantages of FOH have been revealed.
The ubiquinone, coenzyme Q10 (CoQ10; C59H90O4), is an essential cofactor in the aerobic respiratory electron transport chain and is widely distributed in almost all organisms [16, 17]. CoQ10 is composed of a benzoquinone head group and a side chain composed of 10 isoprenoid units, which distinguishes it from other CoQn cognates. CoQ10 is embedded within the hydrophobic domain of the phospholipid bilayer of cytoplasmic membranes, and plays an essential role in removing harmful reactive oxygen species as a redox-active molecule during disulfide bond formation [18, 19]. The clinical benefits of CoQ10 supplementation include the prevention of heart failure as a supplement to 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors [20, 21] and acute myocardial infarction in cardiovascular diseases, as well as positive effects in mitochondrial diseases, certain neurodegenerative diseases, and diabetes [22, 23]. Over the past 2 decades, economical production of CoQ10 using microbial processes has been developed because of the growing demands for CoQ10 in the pharmaceutical and food industry. Some prokaryotic microorganisms, such as Rhodobacter sphaeroides and Agrobacterium tumefaciens, have been well-studied by researchers as natural producers of CoQ10 [20, 24, 25]. In most prokaryotic cells, there are 3 combined pathways involved in CoQ10 biosynthesis. The CoQ10 benzoquinone “head” is synthesized from 4-hydroxybenzoic acid via the shikimate pathway, whereas the isoprenoid side chains are synthesized via isopentenyl pyrophosphate (IPP) synthesis enzymes using 10 IPP molecules derived from pyruvate and glyceraldehyde-3-phosphate from the methylerythritol phosphate (MEP) or mevalonate (MVA) pathways that are only present in eukaryotes and some bacteria. CoQ10 is then assembled by combining the isoprenoid side chain with the benzoquinone head in the ubiquinone pathway [17, 18] (Fig. 1).
The synthesized FPP from IPP and dimethylallyl pyrophosphate (DMAPP) is distributed into different pathways, including lycopene, ubiquinone, or squalene biosynthesis, in prokaryotic microorganisms. R. sphaeroides is utilized for industrial CoQ10 production because a large proportion of FPP can be directed to the ubiquinone synthesis pathway for high-titer CoQ10 production, in addition to the detailed knowledge of the key genes and metabolic pathways involved in CoQ10 biosynthesis [17, 26]. The excess FPP can be used as a precursor for the synthesis of other valuable compounds. In this study, R. sphaeroides GY-2, a high-yield CoQ10 mutant strain obtained from R. sphaeroides 2.4.1 (ATCC 17023) in our previous work [26], was metabolically engineered to directly produce FOH by introducing sesquiterpene synthase or terpene synthase. In order to improve the FPP supplement for FOH biosynthesis, the competitive phytoene synthesis pathway was weakened by efficiently downregulating the psy-coding phytoene synthase (PSY) using RNAi-mediated gene silencing (Fig. 2a). Furthermore, the exogenous genes of phosphatidylglycerophosphatase B (PgpB, GeneID: 945863) from Escherichia coli, terpene synthase (TPS, GeneID: 541974) from Zea mays, and acyclic sesquiterpene synthase (ATPS, GeneID: 100833760) from Brachypodium distachyon were introduced into R. sphaeroides GY-2 for FOH synthesis (Fig. 2b). FOH is synthesized in the cytoplasm and secreted outside the cells, whereas the produced CoQ10 is maintained inside cells and attached to the cell membrane. Therefore, FOH and CoQ10 can theoretically be co-produced simultaneously via separation of supernatant and cell pellet. To the best of our knowledge, this is the first report that is looking to synthesize FOH in R. sphaeroides while attempting to co-produce FOH and CoQ10 at the same time.
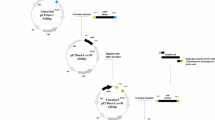
Schematic diagram of the methods used in this work for redirection of carbon flux. a The inducible gene silencing was triggered by expressing a hairpin RNA. After design and induction of the hairpin dsRNA, the dsRNA can be digested by the dicer enzyme in the cell to produce small interference RNA (siRNA) that triggers RNA interference (RNAi) to specifically degrade the endogenous target mRNA. b The short and long fragments of psy are constructed for conformation of the hairpin dsRNA under the control of tac promoter, as well as the exogenous genes coding TPS, PgpB and ATPS
Materials and methods
Chemicals and regents
All the restriction enzymes in this study were purchased from New England Biolabs (Beijing, China). The gel extraction kits and plasmid mini kits were from Omega bio-tek (Guangzhou, China). The standard of farnesol and n-decane are from Sigma-Aldrich Inc., (Milwaukee, Wisconsin, US). The fermentation medium and chemicals were purchased from Beijing Chemical Works (Beijing, China).
Strains and culture conditions
Escherichia coli DH5α and E. coli TOP10 were used for cloning and plasmids construction. E. coli S17-1 is the plasmid donor cells that transport recombinant plasmids into R. sphaeroides strains for bi-parental conjugation. R. sphaeroides GY-2 that is a mutant strain with high yield of CoQ10 was derived from R. sphaeroides 2.4.1. The strains and plasmids used in this study are listed in Table 1. E. coli strains were cultivated at 37 °C in Luria–Bertani (LB) medium supplemented with 50 mg/mL of kanamycin sulfate when necessary. R. sphaeroides strains were grown at 32 °C in the seed culture medium composed of 3.5 g/L (NH4)2SO4, 2 g/L yeast extract, 0.75 g/L corn steep powder, 10 g/L glucose, 2 g/L NaCl, 0.75 g/L K2HPO4, 0.75 g/L KH2PO4, 0.1 g/L FeSO4 and 0.3 g/L MgSO4, 1 g/L nicotinic acid (Amresco Ltd., Solon, US), and 1 g/L biotin (Sigma-Aldrich, St Louis, US) at the pH 6.1–6.3 adjusted by 10 mol/L sodium hydroxide. The fermentation culture medium was composed of 4 g/L (NH4)2SO4, 32.5 g/L glucose, 3.25 g/L NaCl, 1.2 g/L KH2PO4, 1.4 g/L FeSO4 and 9.8 g/L MgSO4, 3.9 g/L corn steep powder with an initial pH 6.0-6.3.
Construction of vectors
The expression plasmid pBBR1MCS2 was modified using pYC6a. The fragment containing the tac promoter and TrrnB terminator was amplified from pYC6a using primers PTR-F and PTR-R. The resulting fragment was digested using SacII and HindIII, and ligated into pBBR1MCS2 to substitute the lac promoter of the original pBBR1MCS2 with tac promoter, resulting in the plasmid pBBR1MCS2-tac. The gene encoding PgpB from E. coli Top 10 was amplified using the primers PgpB-EcoF and PgpB-EcoR listed in Table 1, and ligated with PCR product of pBBR1MCS2 (with primers pBBR1MCS2-F and pBBR1MCS2-R) using Gibson Assembly master mix (New England Biolabs, Beijing, China). The genes encoding ATPS from Brachypodium distachyon and TPS from Zea mays were codon optimized for expression in R. sphaeroides, and synthesized by GenScript Ltd (Nanjing, China). Then codon optimized ATPS and TPS have been ligated into pBBR1MCS2-tac by Gibson Assembly cloning method using the primers ATPS-F, ATPS-R and TPS-F, TPS-R, respectively (Fig. 2b). The protocol of RNAi-mediated gene silencing was followed the previous report [29]. The short (365 bp) and long fragment (462 bp) of psy were amplified from the genome of R. sphaeroides using the primers of PSYSi-F, PSYSi-R and PSYLi-F, PSYLi-R, respectively. Then the short fragment was digested by EcoRI and SacI, whereas the long fragment was digested by EcoRI and SalI. Then ligation was performed by mixing the two digested PCR products (827 bp) with the SacI/SalI digested vector pBBR1MCS2-tac, resulting in the pBBR1MCS2-PSYi that contained the inverted repeat of psy. The genes of PgpB, ATPS and TPS with tac promoter and TrrnB were amplified using the primers PSYitac-F and PSYitac-R, and digested with BamHI/AscI. The three fragments were ligated into the BamHI/AscI digested pBBR1MCS2-PSYi, resulting in the pBBR1MCS2-PSYi-PgpB, pBBR1MCS2-PSYi-ATPS and pBBR1MCS2-PSYi-TPS, respectively (Fig. 2a). Each plasmid and primer sequence was listed in Table 1.
Bi-parental conjugation for transformation
All the pBBR1MCS2-tac derivative expression plasmids were transformed into E. coli S17-1 via electroporation and the resulting strains were used as plasmid donors. The process of conjugation mating was modified and carried out as described in a previous study [30]. Firstly, the donor strain E. coli S17-1 and the recipient R. sphaeroides GY-2 were grown to mid-log phase, harvested by centrifugation (8000g, 6 min) and resuspended in fresh LB medium. Then E. coli S17-1 and R. sphaeroides GY-2 cell suspensions were mixed at the ratios of 1:6 and 1:10, respectively. Afterwards, the culture mixture of the two strains was harvested, transferred to LB agar plate, and incubated for 24–30 h at 32 °C. The resulting strains were harvested by washing with 0.1 mL pre-cooled Sistrom’s minimal medium [31] and spread on LB agar plates containing 200 mg/L K2TeO3 and 50 mg/mL kanamycin. The black kanamycin-resistant colonies were isolated, verified by PCR analysis, and utilized for further experiments. The recombinant R. sphaeroides GY-2 harboring pBBR1MCS2-tac was used as the control.
FOH and CoQ10 production
All the recombinant R. sphaeroides strains were inoculated and grown in the seed culture medium at pH 6.4 and 32 °C, under constant orbital shaking at 200 rpm for 24 h, and then transferred to 500 mL baffled flasks containing 150 mL medium for batch fermentation at the same conditions for 48 h. The method of FOH collection was carried out as described in the previous study [10]. In order to avoid volatile loss of FOH, the fermentation medium was covered with 15% volume of decane to collect FOH. After the batch fermentation, the upper layer of decane phase was collected for FOH extraction. Then the remaining solution was centrifuged for 8 min at 12,000 rpm to collect cells for CoQ10 extraction. The cells were re-suspended using 150 mL PBS solution and 1 mL aliquot of the fermentation broth was harvested, combined 200 μL HCl solution (6 mol/L), and incubated at 65 °C for 20 min. Subsequently, 1 mL acetone and 100 μL of 30% hydrogen peroxide solution were added. After that the mixture was collected, filled with ethanol up to 10 mL, and incubated in an ultrasonic bath at 4 °C for 45 min. Pellets were harvested and centrifuged at 8000g for 10 min. The resulting supernatants were collected and examined for CoQ10 production.
HPLC and GC–MS analysis
The yield of FOH was detected by GC–MS (Agilent Technologies 6890 N Gas Chromatograph combined with 5975C Mass Spectrometry with Triple-Axis Detector) in this study. Samples were injected at a split ratio of 1:5 and were separated using a 19091N-133I HP-INNOWAX chromatographic column (30 m × 0.25 mm × 0.25 μm) with helium at the flow rate of 1.0 mL/min as carrier gas. The initial temperature of the injector was 250 °C and the column oven is set to 60 °C. In the first step of vaporization, the heating rate was 10 °C/min until the temperature rises to 190 °C and held for 2 min. The heating rate in the second step was 20 °C/min to the final temperature of 250 °C and held 5 min. The standard sample of FOH was purchased from Sigma-Aldrich Inc., (Shanghai, China). CoQ10 was determined using an UltiMate TM 3000 HPLC system (Thermo-Fisher Scientific, Waltham, US) equipped with a 250 mm × 4.6 mm × 5 µm, C18-reversed phase column (Thermo-Fisher Scientific, Beijing, China) and a photo-diode array detector. Methanol/isopropyl alcohol at a ratio of 3:1 (v/v) was used as the mobile phase at a flow rate of 0.8 mL/min. CoQ10 can be detected at 275 nm and 254 nm and its concentrations were determined using a standard curve based on an HPLC-grade authentic CoQ10 standard (Sigma-Aldrich, Shanghai, China).
Quantitative real-time RT-PCR analysis
In order to evaluate the effects of RNA interference of the psy gene, quantitative real-time RT-PCR was utilized for analysis of all the PSY RNAi strains. The RNA samples for quantitative real-time RT-PCR analysis were collected from cells grown under the same growth conditions. Total RNA was isolated from all the PSY RNAi R. sphaeroides strains and reversely transcribed using a two-step strategy with reverse transcriptase PCR kit (Qiagen, Shanghai, China). The RT-qPCR reaction was carried out in 25 μL reactions containing 12.5 μL of Universal SYBR Master (ROX), 2.0 μL of properly diluted cDNA from 20–30 ng/mL of cDNA for all genes, 0.5 μL of 50× ROX reference dye II (for error correction between wells), and 0.5 ml of each primer at 10 mM and 9 mL of sterile distilled water. GAPDH gene of R. sphaeroides was selected as control for normalizing expression of the samples. All real-time PCR reactions were performed on the LightCycler® 96 (Roche Diagnostics, US). The fold change of the target cDNA was determined using the value of \(2^{{ - \Delta \Delta {\text{C}}_{\text{t}} }} \left( { - \Delta \Delta {\text{C}}_{\text{t}} = \left[ {{\text{C}}_{{{\text{t}}({\text{target}})}} - {\text{C}}_{{{\text{t}}({\text{ref}})}} } \right]_{\text{sample}} - \left[ {{\text{C}}_{{{\text{t}}({\text{target}})}} - {\text{C}}_{{{\text{t}}({\text{ref}})}} } \right]_{\text{WT}} } \right)\), where Ct represents the threshold cycle [32]. All the values were determined through triplicate experiments and statistical significance was considered significant at P < 0.05.
Results
Introduction of exogenous phosphatases, terpene, and sesquiterpene synthase for FOH production
FOH can be directly synthesized via FPP dephosphorylation in plants and microorganisms. Phosphatases or pyrophosphatases are suitable for FPP catalysis; however, terpene synthase and sesquiterpene synthase are also expected to be catalyst candidates because FOH biosynthesis usually occurs via terpene synthase or sesquiterpene synthase with broad substrate specificities in plants [1, 33]. In this study, the exogenous terpene synthase (TPS) from Zea mays and acyclic sesquiterpene synthase (ATPS) from Brachypodium distachyon, as well as the phosphatidylglycerophosphatase B (PgpB) from E. coli TOP10 were synthesized, codon optimized for expression, and introduced into R. sphaeroides GY-2 for improved FOH production. Figure 3 shows that the introduction of the exogenous genes almost has no effect on the dry cell weight (DCW) of all the strains, except that the biomass accumulation of the R. sphaeroides strain harboring TPS was slightly lower than that of the control strain, R. sphaeroides GY-2. As we expected, FOH and CoQ10 could be co-produced simultaneously from all the metabolically-engineered R. sphaeroides strains (Fig. 4). The results showed that FOH production in the three recombinant R. sphaeroides strains harboring TPS (Rs-TPS), ATPS (Rs-ATPS), and PgpB (Rs-PgpB) was higher than that of the R. sphaeroides strain harboring the plasmid pBBR1MCS2-tac (control) after 48 h of fermentation (Fig. 4a). Thus, it was concluded that FOH could be synthesized using R. sphaeroides GY-2 using its endogenous FOH synthase; however, FOH production could be greatly increased by overexpressing exogenous terpene or sesquiterpene synthase. Rs-TPS, Rs-ATPS, and Rs-PgpB respectively produced 68.2%, 43.4%, and 21.9% higher FOH titers than the control strain. Interestingly, the order of the CoQ10 yield in the 3 recombinant R. sphaeroides strains was exactly opposite to that of FOH production (Fig. 4b). The highest CoQ10 production of 134.5 mg/L was obtained from Rs-PgpB, followed by 133.2 mg/L in Rs-ATPS, and 122.7 mg/L in Rs-TPS, which was lower than the control (131.4 mg/L). As illustrated in Fig. 1, the FOH and CoQ10 synthesis process was based on competition for FPP. When FOH production increased, the FPP used for CoQ10 synthesis correspondingly decreased. Therefore, higher FOH production leads to lower CoQ10 titers in all three recombinant R. sphaeroides strains.
Effect of psy RNA interference on FOH and CoQ10 production
Metabolic flux in microorganisms can be redirected by regulating the key enzymes involved in the pathway. Strategies including overexpression of genes encoding the enzymes for target compound synthesis and disruption of the genes responsible for by-product synthesis or feedback suppression have been developed to improve the titer and yield of the target product [11, 12, 34,35,36]. Here the plasmid pBBR1MCS2-PSYi was constructed for RNA interference of the gene coding phytoene synthase, which converts geranylgeranyl pyrophosphate (GGPP) molecules into phytoene. pBBR1MCS2-PSYi was obtained and transformed into R. sphaeroides GY-2 via bi-parental conjugation. The pre-mRNA was then expressed and spliced to form a self-complementary RNA homologous hairpin comprising of a 365-bp stem and a 62-bp loop by inserting an inverted repeat (IR) sequence. A total of 15 positive psy RNA interference transformants were obtained and confirmed via polymerase chain reaction (PCR) and sequencing using the primers, PSYLi-F and PSYSi-R. Among these 15 positive transformants, 4 strains, designated as PSYi1 to PSYi4, showed significantly decreased transcriptional levels of psy via a primary screening using quantitative real-time RT-PCR analysis, and were selected as the candidate strains. The biomass accumulation results of these candidate strains illustrated that there was no significant difference in the DCW between the 4 psy interference strains and the control R. sphaeroides GY-2 strain (Fig. 3). The DCW of the interference strain cultures could reach up to 14 g/L after 48 h, which was only slightly higher than the control. However, the different psy RNA interference levels in the 4 candidate strains were obtained based on RNAi-mediated gene silencing via pBBR1MCS2-PSYi (Fig. 5a). The expression level of psy in the PSYi2 strain was inhibited by 60%, whereas only 12% inhibition was achieved in PSYi1. This was probably because of the different expression levels of the RNAi or generated siRNA caused due to the different integration sites of the RNAi expression cassette.
FOH and CoQ10 production in all the PSY interference strains was determined using GC–MS and HPLC, respectively (Fig. 5b). The results showed that the final FOH production from all the 4 PSY interference strains was significantly higher than that of the control R. sphaeroides GY-2 strain. The highest FOH yield of 20.98 mg/L was obtained from PSYi4, while R. sphaeroides GY-2 only accumulated 14.2 mg/L, which was 32.3% less than PSYi4. At least more than 20% FOH production was obtained in all the 4 PSY interference strains when compared to the control. The FOH yield from all the PSYi strains was shown to have significantly improved because of the redirection of the carbon flux. The phytoene synthesis pathway was weakened via psy RNA interference, which led to the original flux being directed towards phytoene synthesis and eventually, FOH or CoQ10 synthesis. However, the PSYi2, PSYi3, and PSYi4 CoQ10 titers were not more than 5% higher than the control strain, while the PSYi1 CoQ10 yield was lower compared to the control (Fig. 5b). These results demonstrated that CoQ10 production in the R. sphaeroides strains was almost unaffected by the psy RNA interference and redistributed flux, which suggests that most of the redirected carbon flux from the reduced phytoene synthesis likely contributed to FOH production. Due to the high inhibitory effect of the RNA interference, this RNAi-mediated psy silencing strategy was utilized to further improve the FOH and CoQ10 yields.
Co-production of FOH and CoQ10 from metabolically-engineered R. sphaeroides
In the R. sphaeroides strains, carbon flux competition at the FPP metabolic junction is distributed into different pathways including lycopene synthesis, ubiquinone pathway for CoQ10 production, and squalene biosynthesis. In this study, we opted to reduce the cellular metabolic flux toward squalene biosynthesis in order to enhance the co-production of FOH and CoQ10. Thus, the pBBR1MCS2-PSYi RNA interference plasmid was transformed into Rs-TPS, Rs-ATPS, and Rs-PgpB, and the resulting strains were designated as TPS-PSYi, ATPS-PSYi, and PgpB-PSYi, respectively. Similar to the DCW of the recombinant R. sphaeroides strains in Fig. 2, the TPS-PSYi biomass accumulation was lower than that of the control strain, while no significant difference was found between the ATPS-PSYi, PgpB-PSYi, and control strains (Fig. 6a). When pBBR1MCS2-PSYi was transformed into Rs-TPS, Rs-ATPS, and Rs-PgpB, psy transcription of these 3 R. sphaeroides strains was significantly inhibited (Fig. 6b). More than a 50% decrease in psy transcriptional inhibition was obtained by RNAi in all the 3 strains. Among the 3 RNAi strains, Rs-PgpB was the most inhibited R. sphaeroides strain with only 28.7% transcription remaining when compared to the control. Theoretically, a higher flux through FPP redirects into the ubiquinone and FOH synthesis pathway. Figure 6c demonstrates the FOH production from the metabolically-engineered strains, TPS-PSYi, ATPS-PSYi, and PgpB-PSYi, within 48 h of fermentation. The highest FOH production of 40.45 mg/L was obtained from TPS-PSYi in a batch culture of 48 h, which was twice as high as that of the control strain. Moreover, FOH production from ATPS-PSYi and PgpB-PSYi were 52.8% and 15.6% higher than that of the control, respectively. These results indicate that FOH production can be substantially increased by overexpressing terpene or sesquiterpene synthase coupled with the weakening of the phytoene synthesis pathway via psy RNA interference. Although the highest efficacy of RNA interference was found in PgpB-PSYi, the lowest FOH production was also observed in this strain; this is different from the PSYi2 results, where it was the most inhibited strain but exhibited higher FOH production (Fig. 5). CoQ10 production in TPS-PSYi, ATPS-PSYi and PgpB-PSYi was no more than 120 mg/L, which was lower than that of the control strain, R. sphaeroides GY-2 (Fig. 6d). ATPS-PSYi and TPS-PSYi accumulated the highest and the lowest CoQ10 titers, respectively. CoQ10 production in TPS-PSYi, ATPS-PSYi, and PgpB-PSYi was found to be significantly and respectively lower than the Rs-TPS, Rs-ATPS, and Rs-PgpB overexpression strains. However, no corresponding relationship for the redirecting carbon flux was observed because both FOH and CoQ10 production in ATPS-PSYi was higher than that in PgpB-PSYi.
The metabolically engineered strains TPS-PSYi, ATPS-PSYi and PgpB-PSYi were obtained. a Biomass accumulation of TPS-PSYi, ATPS-PSYi and PgpB-PSYi. b Transcription levels of gene psy of these three R. sphaeroides strains were differently inhibited. c FOH production and d CoQ10 production of TPS-PSYi, ATPS-PSYi and PgpB-PSYi in a 48 h of batch culture
Discussion
Farnesol, an acyclic sesquiterpene alcohol, is found in the essential oils of various plants in nature and is used to treat allergic asthma, gliosis, diabetes, atherosclerosis, and hyperlipidemia [8]. Exogenous FOH has been reported to regulate inflammatory responses and exhibit anti-cancerous and anti-inflammatory effects [8, 9]. FOH, extracted from natural plants, has been used in chemoprevention due to its relatively low toxicity as compared to its synthetic counterparts; however, its extraction cannot meet large scale needs because limited amounts of FOH content in plant oil leads to high extraction costs. There is a growing interest in engineering terpene and sesquiterpene production for large-scale fermentation using microorganisms to circumvent the limited availability of these valuable compounds with low natural content [12]. In this study, we firstly produced FOH from a metabolically-engineered R. sphaeroides mutant strain GY-2 that already provides high CoQ10 yields, which is also a valued compound with many clinical benefits [16]. FOH and CoQ10 can be co-produced simultaneously and separated easily from one another because the synthesized FOH is secreted outside cells, whereas CoQ10 is produced and maintained inside the cells. Although FOH can be synthesized from R. sphaeroides GY-2, it is almost impossible to obtain more than 15 mg/L of FOH due to the low enzymatic activities of endogenous FOH synthase or pyrophosphorylase in the control strain. FOH production was significantly increased by overexpressing PgpB from Escherichia coli, TPS from Zea mays, and ATPS from Brachypodium distachyon in R. sphaeroides GY-2. However, CoQ10 production decreased correspondingly due to the enhanced carbon flux directed toward FOH synthesis. These results suggested that an improved flux, including the portion originally used for CoQ10 production, was redirected toward FOH synthesis through metabolic modification.
On the other hand, the reaction catalyzed by PSY, which was the first committed step of the carotenoid synthesis, was considered to be an important control point in regulating the carbon flux through the carotenoid synthesis pathway [37]. Carotenoids in microorganisms are considered to be biological antioxidants against reactive oxygen species that relieve the damage caused by oxidative DNA-damaging agents [38]. RNA interference mediated by the expression of short hairpin RNAs (shRNAs) is a powerful tool in efficiently suppressing target genes. Thus, RNA interference-mediated silencing, rather than disruption, of psy was carried out in this work to weaken the phytoene synthesis pathway so that the carbon flux directed towards FOH and CoQ10 could be further improved. Phytoene production was decreased by 26.3% when compared to the control in the PSY interference strains (Additional file 1: Fig. S1), resulting in improved FOH production.
There is probably not much flux being directed into the functional carotenogenic pathway because only a small amount of carotenoid can be found in many organisms [39]. However, improved FOH production was obtained in all four PSY interference strains when compared to the control strain, whereas CoQ10 production in these strains was almost unaffected (Fig. 5). In fact, conditional inhibition of the specific pathway after sufficient bacterial growth has been shown to redirect the excess flux toward a desired bio-production pathway [40]. The results suggested that most of the redirected excess carbon flux caused by the interruption of phytoene synthesis likely contributed to FOH production. An alternative explanation to the results is that the flux redirected towards improved FOH production was far more significant than that toward CoQ10 production because the same amount of redirected flux from the reduced phytoene synthesis contributed to almost 4 times more FOH than CoQ10 due to the different carbon contents of their molecules. Furthermore, FOH production was greatly improved by the three metabolically-engineered strains combined with the terpene or sesquiterpene synthase overexpression and psy RNA interference (Fig. 6c). CoQ10 production in these strains was also significantly decreased and even lower than that of the control strain, R. sphaeroides GY-2 (Fig. 6d). This confirmed the previous suggestion that the improved flux that was originally involved in CoQ10 production was being redirected towards FOH synthesis through metabolic modification. It appears that the carbon flux balance can be disturbed and redirected when a related metabolic pathway changes. In this study, a generated carbon flux derived from multiple sources at the FPP metabolic junction was redistributed towards FOH synthesis by introducing exogenous terpene or sesquiterpene synthase. The TPS-PSYi and ATPS-PSYi strains exhibited potential industrial application prospects due to their ability to simultaneously co-produce high levels of FOH and CoQ10. However, the highest FOH titer of 40.45 mg/L obtained from TPS-PSYi in this study was still lower than the previous reports [10, 11] due to lack of MVA pathway in R. sphaeroides. Despite this disadvantage, the strategy in this work for effective metabolic flux redirection is convenient and economical for the possible commercial co-production of FOH and CoQ10. Further improvements in FOH and CoQ10 production can be possibly accomplished through metabolic engineering for more available carbon flux redirected towards the desired products.
Conclusions
In this study, co-production of FOH and CoQ10 was performed by introducing three different exogenous terpenes or sesquiterpene synthases combined with psy RNA interference in R. sphaeroides GY-2. It is noteworthy that FOH production was greatly enhanced due to redirected flux derived from the carotenoid and CoQ10 synthesis pathways. This study confirms the effectiveness of psy RNA interference in redirecting carbon flux, responsible for phytoene synthesis, towards FOH production. To the best of our knowledge, this is the first reported instance of simultaneous FOH and CoQ10 co-production in R. sphaeroides using the metabolic engineering strategy. This strategy can broaden applications in other organisms for further improvements in desired products. Therefore, the two strains, TPS-PSYi and ATPS-PSYi, developed in this study provide a viable approach using strain engineering for the industrial production of FOH and CoQ10.
Availability of data and materials
The datasets used and/or analyzed during the current study are included in this article and available from the corresponding author on reasonable request.
Abbreviations
- PSY:
-
phytoene synthase
- PPFPP:
-
prephytoene diphosphate
- GGPP:
-
geranylgeranyl-PP
- UbiA:
-
4HBA-polyprenyltransferase
- UbiD, B, E, F, G, H:
-
quinoid ring modification enzymes
- DMAPP:
-
dimethylallyl-PP
- IPP:
-
isopentenyl-PP
- GPP:
-
geranyl-PP
- FPP:
-
(E,E)-farnesyl-PP
- GGPP:
-
geranylgeranyl-PP
- IDI:
-
isopentenyl-diphosphate delta-isomerase
- DXR:
-
1-deoxy-d-xylulose-5-phosphate reductoisomerase
- ispD:
-
4-diphosphocytidyl-2-C-methyl-d-erythritol synthase (IspD)
- ispE:
-
4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (IspE)
- ispF:
-
2-C-methyl-d-erythritol-2,4-cyclodiphosphate synthase (IspF)
- ispG:
-
4-hydroxy-3-methyl-but-2-enyl pyrophosphate synthase (IspG)
- ispH:
-
4-hydroxy-3-methyl- but-2-enyl pyrophosphate reductase (IspH)
- MvaE:
-
bifunctional acetoacetyl-CoA thiolase and HMG-CoA reductase
- MvaS:
-
hMG-CoA synthase
- MvaK1:
-
mevalonate kinase
- MvaK2:
-
phosphomevalonate kinase
- HMG-CoA:
-
hydroxymethylglutaryl-CoA
- MVA:
-
mevalonate
- MVA-P:
-
mevalonate 5-phosphate
- MVA-PP:
-
mevalonate pyrophosphate
References
Dewick PM. The biosynthesis of C5–C25 terpenoid compounds. Nat Prod Rep. 2002;19:181–222.
Bakkali F, Averbeck S, Averbeck D, Idaomar M. Biological effects of essential oils—a review. Food Chem Toxicol. 2008;46:446–75.
Lorek J, Poggeler S, Weide MR, Breves R, Bockmuhl DP. Influence of farnesol on the morphogenesis of Aspergillus niger. J Basic Microbiol. 2008;48:99–103.
Shea JM, Del Poeta M. Lipid signaling in pathogenic fungi. Curr Opin Microbiol. 2006;9:352–8.
Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67:2982–92.
Joo JH, Jetten AM. Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett. 2010;287:123–35.
Shirtliff ME, Krom BP, Meijering RA, Peters BM, Zhu J, Scheper MA, Harris ML, Jabra-Rizk MA. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother. 2009;53:2392–401.
Lee JH, Kim C, Kim SH, Sethi G, Ahn KS. Farnesol inhibits tumor growth and enhances the anticancer effects of bortezomib in multiple myeloma xenograft mouse model through the modulation of STAT3 signaling pathway. Cancer Lett. 2015;360:280–93.
Jung YY, Hwang ST, Sethi G, Fan L, Arfuso F, Ahn KS. Potential anti-inflammatory and anti-cancer properties of farnesol. Molecules. 2018;23:2827.
Wang C, Yoon SH, Shah AA, Chung YR, Kim JY, Choi ES, Keasling JD, Kim SW. Farnesol production from Escherichia coli by harnessing the exogenous mevalonate pathway. Biotechnol Bioeng. 2010;107:421–9.
Wang C, Park JE, Choi ES, Kim SW. Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway—application of PgpB and YbjG phosphatases. Biotechnol J. 2016;11:1291–7.
Takahashi S, Yeo Y, Greenhagen BT, McMullin T, Song L, Maurina-Brunker J, Rosson R, Noel JP, Chappell J. Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol Bioeng. 2007;97:170–81.
Asadollahi MA, Maury J, Møller K, Nielsen KF, Schalk M, Clark A, Nielsen J. Production of plant sesquiterpenes in Saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol Bioeng. 2008;99:666–77.
Muramatsu M, Ohto C, Obata S, Sakuradani E, Shimizu S. Alkaline pH enhances farnesol production by Saccharomyces cerevisiae. J Biosci Bioeng. 2009;108:52–5.
Hornby JM, Nickerson KW. Enhanced production of farnesol by Candida albicans treated with four azoles. Antimicrob Agents Chemother. 2004;48:2305–7.
Bader MW, Xie T, Yu CA, Bardwell JC. Disulfide bonds are generated by quinone reduction. J Biol Chem. 2000;275:26082–8.
Cluis CP, Ekins A, Narcross L, Jiang H, Gold ND, Burja AM, Martin VJ. Identification of bottlenecks in Escherichia coli engineered for the production of CoQ10. Metab Eng. 2011;13:733–44.
Crane FL. Biochemical functions of coenzyme Q10. J Am Coll Nutr. 2001;20:591–8.
Zahiri HS, Yoon SH, Keasling JD, Lee SH, Won Kim S, Yoon SC, Shin YC. Coenzyme Q10 production in recombinant Escherichia coli strains engineered with a heterologousdecaprenyl diphosphate synthase gene and foreign mevalonate pathway. Metab Eng. 2006;8:406–16.
Choi G, Kim Y, Seo J, Ryu Y. Restricted electron flux increases coenzyme Q10 production in Agrobacterium tumefaciens ATCC4452. Process Biochem. 2005;40:3225–9.
Folkers K, Langsjoen P, Willis R, Richardson P, Xia LJ, Ye CQ, Tamagawa H. Lovastatin decreases coenzyme Q levels in humans. Proc Natl Acad Sci USA. 1990;87:8931–4.
Hodgson JM, Watts GF, Playford DA, Burke V, Croft KD. Coenzyme Q10 improves blood pressure and glycaemic control: a controlled trial in subjects with type 2 diabetes. Eur J Clin Nutr. 2002;56:1137–42.
Bhagavan HN, Chopra RK. Potential role of ubiquinone (coenzyme Q10) in pediatric cardiomyopathy. Clin Nutr. 2005;24:331–8.
Kien NB, Kong IS, Lee MG, Kim JK. Coenzyme Q10 production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J Ind Microbiol Biotechnol. 2010;37:521–9.
Lu W, Ye L, Xu H, Xie W, Gu J, Yu H. Enhanced production of coenzyme Q10 by self-regulating the engineered MEP pathway in Rhodobacter sphaeroides. Biotechnol Bioeng. 2014;111:761–9.
Qi F, Zou L, Jiang X, Cai S, Zhang M, Zhao X, Huang J. Integration of heterologous 4-hydroxybenzoic acid transport proteins in Rhodobacter sphaeroides for enhancement of coenzyme Q10 production. RSC Adv. 2017;7:17346–52.
Park YC, Kim SJ, Choi JH, Lee WH, Park KM, Kawamukai M, Ryu YW, Seo JH. Batch and fed-batch production of coenzyme Q10 in recombinant Escherichia coli containing the decaprenyl diphosphate synthase gene from Gluconobacter suboxydans. Appl Microbiol Biotechnol. 2005;67:192–6.
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6.
Qin LN, Cai FR, Dong XR, Huang ZB, Tao Y, Huang JZ, Dong ZY. Improved production of heterologous lipase in Trichoderma reesei by RNAi mediated genesilencing of an endogenic highly expressed gene. Bioresour Technol. 2012;109:116–22.
Porter SL, Wadhams GH, Armitage JP. In vivo and in vitro analysis of the Rhodobacter sphaeroides chemotaxis signaling complexes. Methods Enzymol. 2007;423:392–413.
Moore MD, Kaplan S. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1505–14.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods. 2001;25:402–8.
Wang C, Kim JY, Choi ES, Kim SW. Microbial production of farnesol (FOH): current states and beyond. Process Biochem. 2011;46:1221–9.
Wang W, Liu X, Lu X. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol Biofuels. 2013;6:69.
Wang CW, Oh MK, Liao JC. Engineere disoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol Bioeng. 1999;62:235–41.
Chen Z, Liu D. Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol Biofuels. 2016;9:205.
Sandmann G, Römer S, Fraser PD. Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab Eng. 2006;8:291–302.
Zhang L, Yang Q, Luo X, Fang C, Zhang Q, Tang Y. Knockout of crtB or crtI gene blocks the carotenoid biosynthetic pathway in Deinococcus radiodurans R1 and influences its resistance to oxidative DNA-damaging agents due to change of free radicals scavenging ability. Arch Microbiol. 2007;188:411–9.
Sandmann G. Carotenoid biosynthesis in microorganisms and plants. EJB Reviews. 1994;223:7–24.
Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng. 2014;23:175–84.
Acknowledgements
Authors are grateful to Prof. Lina Qin for her important technical support and critical discussion in this work.
Funding
This work was financially supported by National Natural Science Foundation of China (21406130, 31800060, 31741002), Natural Science Foundation of Fujian Province of China (2016J01148, 2016J01147) and College of Life Sciences, Fujian Normal University (FZSKB2018002).
Author information
Authors and Affiliations
Contributions
XC and MX performed all of the experiments and data processing. They contributed equally to this work. MZ and RH performed parts of the experiments and data processing. XZ and FQ designed the experiments and prepared manuscript. JH contributed to manuscript preparation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file

Additional file 1: Fig. S1.
Phytoene production of the four strains with RNAi mediated silencing of the gene psy. R. sphaeroides GY-2 was used as the control.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, X., Jiang, X., Xu, M. et al. Co-production of farnesol and coenzyme Q10 from metabolically engineered Rhodobacter sphaeroides. Microb Cell Fact 18, 98 (2019). https://doi.org/10.1186/s12934-019-1145-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-019-1145-6