Abstract
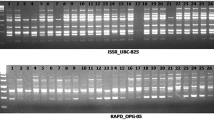
Access to genetic diversity is essential for any progress in adapting linseed (Linum usitatissimum subsp. usitatissimum L.) cultivation to changing environmental conditions or to the changing market needs. An attempt has been made in the present study to assess genetic diversity in 96 genotypes of linseed including varieties, landraces and exotic material. A total of 38 SSR primers amplified 153 alleles with 4.0 alleles per marker locus. The number of alleles ranged from 2 to 15 and the observed polymorphism ranged from 50 to 100%. Average genetic dissimilarity ranged from 2 to 50%. In order to analyze the efficiency for unambiguous identification of linseed germplasm, various statistical measures, viz., number of genotyping patterns, polymorphism information content, resolving power, discrimination power, probability of identity and probability of random identity, identified a set comprising of primers LU7, LU27, LU25, LU20 and LU31 (or LU637) for DNA fingerprinting of linseed germplasm. UPGMA cluster analysis showed that all genotypes could be grouped into four main clusters. Cluster 2 was the largest consisting of mainly landraces, whereas, Cluster 4 was the smallest. Cluster 1 consisted of mainly the released cultivars. Cluster 3 and Cluster 4 were smaller clusters and consisted of exotic genotypes. Principal co-ordinate analysis further substantiated the UPGMA clustering patterns of the observed genetic relationship. To explain 70–80% variability, 17–23 PCOs were needed, whereas 70 components were needed to explain the whole variability in the linseed material under study. Analysis of molecular variance indicated that most of the genetic variation is owing to the individuals within single population, whereas grouping of linseed material into varieties, landraces and exotics accounted for nearly 10% of the total genetic variation. The utility of SSR markers in diversity assessment and cultivar identification is discussed.


Similar content being viewed by others

Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- CTAB:
-
Cetyltrymethylammonium bromide
- ISSR:
-
Inter-simple sequence repeats
- RAPD:
-
Random amplified polymorphic DNA
- RFLP:
-
Restriction fragment length polymorphism
- SRAP:
-
Sequence related amplified polymorphism
References
Adugna W, Labuschange MT, Viljoen CD (2006) The use of morphological and AFLP markers in diversity analysis of linseed. Biodivers Conserv 15:3193–3205
Anderson JA, Churchill GA, Autroque JE, Tanksley SD, Swells ME (1993) Optimising selection for plant linkage map. Genome 36:181–186
Anonymous (2015) E-book of Agricultural Statistics at a glance 2015, Directorate of Economics and Statistics, Department, Agriculture, Cooperation & Famers Welfare, Ministry of Agriculture & Farmers Welfare, Government of India. http://eands.dacnet.nic.in
Bhat TM, Kudesia R, Srivastav MK, Sidiqqui S (2011) Evaluation of variability in five linseed (Linum usitatissimum L.) genotypes using agromorphological characters and RAPD analysis. South Asian J Exp Biol 1:43–47
Botstein D, White RL, Skolnick M, Davis RM (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Cloutier S, Niu Z, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119:53–63
Deng X, Long S, He D, Li X, Wang Y, Liu J, Chen X (2010) Development and characterization of polymorphic microsatellite markers in Linum usitatissimum. J Plant Res 123:119–123
Deng X, Long S, He D, Li X, Wang Y, Hao D, Qiu C, Chen X (2011) Isolation and characterization of polymorphic microsatellite markers from flax (Linum usitatissimum L.). Afr J Biotechnol 10:734–739
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fu YB (2005) Geographic patterns of RAPD variation in cultivated flax. Crop Sci 45:1084–1091
Fu YB (2006) Redundancy and distinctiveness in flax germplasm as revealed by RAPD dissimilarity. Plant Genet Resour 4:117–124
Fu YB, Peterson GW (2010) Characterization of expressed sequence tag-derived simple sequence repeat markers for 17 Linum species. Botany 88:537–543
Fu YB, Diederichsen A, Richards KW, Peterson G (2002) Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPD. Genet Resour Crop Evol 49(2):167–174
Green AG, Chen Y, Singh SP, Dribnenki JCP (2008) Flax. In: Kole C, Hall TC (eds) Compendium of transgenic crop plants: transgenic oilseed crops. Blackwell Publishing Ltd, Oxford, pp 199–226
Gupta PS, Srivastava MK, Gupta A (2010) Genetic diversity and fingerprinting of flax cultivars of India using RAPD-PCR markers. Guangxi Agric Sci 41(3):201–206
Lemesh V (2009) Identification of flax genotypes using RAPD and SSR markers. Vagos 82:12–15
Li M (2011) Genetic diversity and relationship of flax germplasm as revealed by AFLP analysis. Crop genetics and breeding germplasm resources. Acta Agron Sin 37:635–640
Li D, Yadong Z, Chao L, Tu S, Li M (2009) Optimization of SRAP-PCR reaction system in flax. J Northeast Agric Univ 16:17–22
Muravenko OV, Bolsheva NL, Yurkevich OY, Nosova IV, Rachinskaya OA, Samatadze TE, Zelenin AV (2010) Karyogenomics of species of the genus Linum L., Russian. J Genet 46(10):1182–1185
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Rajwade AV, Arora RS, Kadoo NY, Harsulkar AM, Ghorpade PB, Gupta VS (2010) Relatedness of Indian flax genotypes (Linum usitatissimum L.): an inter-simple sequence repeat (ISSR) primer assay. Mol Biotechnol 45:161–170
Rana MK, Munjal DV, Singh S, Bhat KV (2005) Utility of P450-based-analogue (PBAs) markers for diversity analysis of linseed (Linum usitatissimum L.) cultivars. Indian J Plant Genet Resour 18(2):194–199
Rohlf FJ (2002) NTSYS-pc Numerical taxonomy and multivariate analysis system. Version 2.1a. Exeter Publ. Setauket, New York
Roose-Amsaleg C, Cariou-Pham E, Vautrin D, Tavernier R, Solignac M (2006) Polymorphic microsatellite loci in Linum usitatissimum. Mol Ecol Notes 6:796–799
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci 81:8014–8018
Singh PK, Akram M, Srivastava RL (2009) Genetic diversity in linseed (Linum usitatissimum) cultivars based on RAPD analysis. Indian J Agric Sci 79(12):1046–1049
Smykal P, Bacova K, Kalender R, Corander J, Schulman AH, Pavelek M (2011) Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor Appl Genet 122:1385–1397
Soto-Cerda BJ, Carrasco RA, Aravena GA, Urbina HA, Navarro CS (2011a) Identifying novel polymorphic microsatellites from cultivated flax (Linum usitatissimum L.) following data mining. Plant Mol Biol Report 29:753–759
Soto-Cerda BJ, Urbina-Saavedra H, Navarro C, Mora-Ortega P (2011b) Characterization of novel genic SSR markers in Linum usitatissimum (L.) and their transferability across eleven Linum species. Electron J Biotechnol 14(2):6
Soto-Cerda BJ, Maureira-Butler I, Muñoz G, Rupayan A, Cloutier S (2012) SSR based population structure, molecular diversity and linkage disequilibrium analysis of a collection of flax (Linum usitatissimum L.) varying for mucilage seed-coat content. Mol Breed 30:875–888
Soto-Cerda BJ, Diederichsen A, Ragupathy R, Cloutier S (2013) Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol 13:78
Soto-Cerda BJ, Westermeyer F, Iñiguez-Luy F, Muñoz G, Montenegro A, Cloutier S (2014a) Assessing the agronomic potential of linseed genotypes by multivariate analyses and association mapping of agronomic traits. Euphytica 196:35–49
Soto-Cerda BJ, Duguid S, Booker H, Rowland G, Diederichsen A, Cloutier S (2014b) Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection. Theor Appl Genet 127(4):881–896
Tessier C, David J, This P, Boursiquot JM, Charrier A (1999) Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet 98:171–177
Uysal H, Fu YB, Kurt O, Peterson GW, Diederichsen A, Kusters P (2010) Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57:1109–1119
Vavilov NI (1951) The origin, variation, immunity and breeding of cultivated plants. Chron Bot 13:1–366
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396
Wiesner I, Wiesnerova D, Tejklova E (2001) Effect of anchor and core sequence in microsatellite primers on flax fingerprinting patterns. J Agric Sci 137:37–44
Wiesnerova D, Wiesner I (2004) ISSR-based clustering of cultivated flax germplasm is statistically correlated to thousand seed mass. Mol Biotechnol 26:207–214
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 29:157
Acknowledgements
We greatly acknowledge financial support from the Indian Council of Agricultural Research (ICAR). Director, NBPGR and Officer-In-Charge, Division of Genomic Resources, NBPGR are also acknowledged for providing laboratory facilities for carrying out this work. Thanks are also due to the Council of Scientific and Industrial Research (CSIR) for awarding Senior Research Associateship to Ms. Sonika Singh.
Funding
This study was partly funded by Council of Scientific and Industrial Research (CSIR) to Sonika Singh in terms of Senior Research Associateship vide CSIR Letter No. 13(8547-A)/2012/pool dated 17/04/2012.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rana, M.K., Singh, S. Assessment of genetic diversity and DNA profiling of linseed (Linum usitatissimum subsp. usitatissimum L.) germplasm using SSR markers. J. Plant Biochem. Biotechnol. 26, 293–301 (2017). https://doi.org/10.1007/s13562-016-0391-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-016-0391-5