Abstract
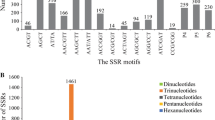
A set of 146,611 expressed sequence tags (ESTs) were generated from 10 flax cDNA libraries. After assembly, a total of 11,166 contigs and 11,896 singletons were mined for the presence of putative simple sequence repeats (SSRs) and yielded 806 (3.5%) non-redundant sequences which contained 851 putative SSRs. This is equivalent to one EST-SSR per 16.5 kb of sequence. Trinucleotide motifs were the most abundant (76.9%), followed by dinucleotides (13.9%). Tetra-, penta- and hexanucleotide motifs represented <10% of the SSRs identified. A total of 83 SSR motifs were identified. Motif (TTC/GAA)n was the most abundant (10.2%) followed by (CTT/AAG)n (8.7%), (TCT/AGA)n (8.6%), (CT/AG)n (6.7%) and (TC/GA)n (5.3%). A total of 662 primer pairs were designed, of which 610 primer pairs yielded amplicons in a set of 23 flax accessions. Polymorphism between the accessions was found for 248 primer pairs which detected a total of 275 EST-SSR loci. Two to seven alleles were detected per marker. The polymorphism information content value for these markers ranged from 0.08 to 0.82 and averaged 0.35. The 635 alleles detected by the 275 polymorphic EST-SSRs were used to study the genetic relationship of 23 flax accessions. Four major clusters and two singletons were observed. Sub-clusters within the main clusters correlated with the pedigree relationships amongst accessions. The EST-SSRs developed herein represent the first large-scale development of SSR markers in flax. They have potential to be used for the development of genetic and physical maps, quantitative trait loci mapping, genetic diversity studies, association mapping and fingerprinting cultivars for example.




Similar content being viewed by others
References
Adugna W, Viljoen CD, Labuschagne MT (2005) Analysis of genetic diversity in linseed using AFLP markers. Sinet Ethiop J Sci 28:41–50
Adugna W, Labuschagne MT, Viljoen CD (2006) The use of morphological and AFLP markers in diversity analysis of linseed. Biodivers Conserv 15:3193–3205
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Benham J, Jeung J-U, Jasieniuk M, Kanazin V, Blake T (1999) Genographer: a graphical tool for automated fluorescent AFLP and microsatellite analysis. J Agric Genom 4. http://wheat.pw.usda.gov/jag/
Benneth MD, Leitch IJ (2005) Nuclear DNA amounts in angiosperms—progress, problems and prospects. Ann Bot 95:45–90
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156:847–854
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
Diederichsen A (2001) Comparison of genetic diversity of flax (Linum usitatissimum L.) between Canadian cultivars and a world collection. Plant Breed 120:360–362
Diederichsen A (2007) Ex situ collections of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genet Resour Crop Evol 54:661–678
Diederichsen A, Hammer K (1995) Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet Res Crop Evol 42:263–272
Diederichsen A, Raney JP (2006) Seed colour, seed weight and seed oil content in Linum usitatissimum accessions held by Plant Gene Resources of Canada. Plant Breed 125:372–377
Diederichsen A, Raney JP, Duguid SD (2006) Variation of mucilage in flax seed and its relationship with other seed characters. Crop Sci 46:365–371
Dillman AC (1953) Classification of flax varieties, 1946. US Dept. of Agriculture, 1953. SERIES INFORMATION: Technical bulletin/United States Department of Agriculture; no. 1064. PUBLISHER: US Dept of Agriculture, Washington
Dribnenki JCP, Green AG (1995) Linola ‘947’ low linolenic acid flax. Can J Plant Sci 75:201–202
Dribnenki JCP, McEachern SF, Green AG, Kenaschuk EO, Rashid KY (1999) Linola ‘1084’ low-linolenic acid flax. Can J Plant Sci 79:607–609
Dribnenki JCP, McEachern SF, Chen Y, Green AG, Rashid KY (2003) LinolaTM 2047 low linolenic acid flax. Can J Plant Sci 83:81–83
Dribnenki JCP, McEachern SF, Chen Y, Green AG, Rashid KY (2004) 2090 low linolenic acid flax. Can J Plant Sci 84:797–799
Dribnenki JCP, McEachern SF, Chen Y, Green AG, Rashid KY (2005) 2126 low linolenic flax. Can J Plant Sci 85:155–157
Dribnenki JCP, McEachern SF, Chen Y, Green AG, Rashid KY (2007) 2149 solin (low linolenic flax). Can J Plant Sci 87:297–299
Duguid SD, Kenaschuk EO, Rashid KY (2003a) Hanley flax. Can J Plant Sci 83:85–87
Duguid SD, Kenaschuk EO, Rashid KY (2003b) Lightning flax. Can J Plant Sci 83:89–91
Duguid SD, Kenaschuk EO, Rashid KY (2003c) Macbeth flax. Can J Plant Sci 83:803–805
Duguid SD, Kenaschuk EO, Rashid KY (2004) Prairie blue flax. Can J Plant Sci 84:801–803
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407
Everaert I, Riek JD, Loose MD, Waes JV, Bockstaele EV (2001) Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds 14:69–87
Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor Appl Genet 111:456–466
Fraser LG, Harvey CF, Crowhurst RN, De Silva HN (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016
Fu Y, Peterson G, Diederichsen A, Richards KW (2002) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Res Crop Evol 49:253–259
Fu Y, Rowland GG, Duguid SD, Richards KW (2003a) RAPD analysis of 54 North American flax cultivars. Crop Sci 43:1510–1515
Fu YB, Guerin S, Peterson GW, Diederichsen A, Rowland GG, Richards KW (2003b) RAPD analysis of genetic variability of regenerated seeds in the Canadian flax cultivar CDC Normandy. Seed Sci Technol 31:207–211
Fu YB, Peterson GW, Yu JK, Gao L, Jia J, Richards KW (2006) Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by EST-derived SSR markers. Theor Appl Genet 112:1239–1247
Gong L, Stift G, Kofler R, Pachner M, Lelley T (2008) Microsatellites for the genus Cucurbita and an SSR-based genetic linkage map of Cucurbita pepo L. Theor Appl Genet 117:37–48
Gorman MB, Cullis CA, Aldridge N (1993) Genetic and linkage analysis of isozyme polymorphisms in flax. J Hered 84:73–78
Han ZG, Guo WZ, Song XL, Zhang TZ (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Genomics 272:308–327
Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Hisano H, Sato S, Isobe S, Sasamoto S, Wada T, Matsuno A, Fujishiro T, Yamada M, Nakayama S, Nakamura Y, Watanabe S, Harada K, Tabata S (2007) Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res 14:271–281
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, Somers DJ, Brown PD (2006) Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor Appl Genet 113:753–766
Kenaschuk EO, Rashid KY (1993) AC Linora flax. Can J Plant Sci 73:839–841
Kenaschuk EO, Rashid KY (1994) AC McDuff flax. Can J Plant Sci 74:815–816
Kenaschuk EO, Rashid KY (1998) AC Watson flax. Can J Plant Sci 78:465–466
Kenaschuk EO, Rashid KY (1999) Nouveau cultivar de lin AC Carnduff. Can J Plant Sci 79:373–374
Kenaschuk EO, Rashid KY, Gubbels GH (1996) AC Emerson flax. Can J Plant Sci 76:483–485
Krulickova K, Posvec Z, Griga M (2002) Identification of flax and linseed cultivars by isozyme markers. Biol Plant 45:327–336
La Rota M, Kantety RV, Yu JK, Sorrells ME (2005) Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics 6:23
McBreen K, Lockhart PJ, McLenachan PA, Scheele S, Robertson AW (2003) The use of molecular techniques to resolve relationships among traditional weaving cultivars of Phormium. NZ J Bot 41:301–310
Mian MA, Saha MC, Hopkins AA, Wang ZY (2005) Use of tall fescue EST-SSR markers in phylogenetic analysis of cool-season forage grasses. Genome 48:637–647
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Oh TJ, Gorman M, Cullis CA (2000) RFLP and RAPD mapping in flax (Linum usitatissimum). Theor Appl Genet 101:590–593
Park YH, Alabady MS, Ulloa M, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, el-Shihy OM, Cantrell RG (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol Genet Genom 274:428–441
Peng JH, Lapitan NL (2005) Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct Integr Genomics 5:80–96
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Roose Amsaleg C, Cariou Pham E, Vautrin D, Tavernier R, Solignac M (2006) Polymorphic microsatellite loci in Linum usitatissimum. Mol Ecol Notes 6:796–799
Rowland GG, Hormis YA, Rashid KY (2002) CDC Bethune flax. Can J Plant Sci 82:101–102
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Rungis D, Berube Y, Zhang J, Ralph S, Ritland CE, Ellis BE, Douglas C, Bohlmann J, Ritland K (2004) Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor Appl Genet 109:1283–1294
Saeidi G (2008) Genetic variation and heritability for germination, seed vigour and field emergence in brown and yellow-seeded genotypes of flax. Int J Plant Prod 2:15–22
Saha MC, Cooper JD, Mian MA, Chekhovskiy K, May GD (2006) Tall fescue genomic SSR markers: development and transferability across multiple grass species. Theor Appl Genet 113:1449–1458
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Spielmeyer W, Green AG, Bittisnich D, Mendham N, Lagudah ES (1998) Identification of quantitative trait loci contributing to Fusarium wilt resistance on an AFLP linkage map of flax (Linum usitatissimum). Theor Appl Genet 97:633–641
Stegnii VN, Chudinova YV, Salina EA (2000) RAPD analysis of flax (Linum usitatissimum L.) varieties and hybrids of various productivity. Genetika Moskva 36:1370–1373
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Tyson H, Fieldes MA, Cheung C, Starobin J (1985) Isozyme relative mobility changes relative to leaf position; apparently smooth trends and some implications. Biochem Genet 23:641–654
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Varshney RK, Grosse I, Hahnel U, Siefken R, Prasad M, Stein N, Langridge P, Altschmied L, Graner A (2006) Genetic mapping and BAC assignment of EST-derived SSR markers shows non-uniform distribution of genes in the barley genome. Theor Appl Genet 113:239–250
Wiesner I, Wiesnerova D, Tejklova E (2001) Effect of anchor and core sequence in microsatellite primers on flax fingerprinting patterns. J Agric Sci 137:37–44
Xu Y, Ma RC, Xie H, Liu JT, Cao MQ (2004) Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 47:1091–1104
Yu JK, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Zhang LY, Ravel C, Bernard M, Balfourier F, Leroy P, Feuillet C, Sourdille P (2006) Transferable bread wheat EST-SSRs can be useful for phylogenetic studies among the Triticeae species. Theor Appl Genet 113:407–418
Zohary D (1999) Monophyletic vs. polyphyletic origin of the crops on which agriculture was founded in the Near East. Genetic Res Crop Evol 46:133–142
Acknowledgments
The authors are grateful to Andrzej Walichnowski for manuscript review, Joanne Schiavoni for manuscript preparation and Mike Shillinglaw for the preparation of figures. Technical assistance of Laura Marginet, Natalie Middlestead, Mira Popovic, Elsa Reimer and Andrzej Walichnowski is also acknowledged. Travis Banks provided most helpful bioinformatics expertise throughout the project. This work was financially supported by AAFC A-base project for high-value flax development. This is AAFC contribution number 1972.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Xue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cloutier, S., Niu, Z., Datla, R. et al. Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119, 53–63 (2009). https://doi.org/10.1007/s00122-009-1016-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1016-3