Abstract
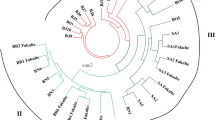
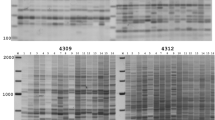
Retrotransposon segments were characterized and inter-retrotransposon amplified polymorphism (IRAP) markers developed for cultivated flax (Linum usitatissimum L.) and the Linum genus. Over 75 distinct long terminal repeat retrotransposon segments were cloned, the first set for Linum, and specific primers designed for them. IRAP was then used to evaluate genetic diversity among 708 accessions of cultivated flax comprising 143 landraces, 387 varieties, and 178 breeding lines. These included both traditional and modern, oil (86), fiber (351), and combined-use (271) accessions, originating from 36 countries, and 10 wild Linum species. The set of 10 most polymorphic primers yielded 141 reproducible informative data points per accession, with 52% polymorphism and a 0.34 Shannon diversity index. The maximal genetic diversity was detected among wild Linum species (100% IRAP polymorphism and 0.57 Jaccard similarity), while diversity within cultivated germplasm decreased from landraces (58%, 0.63) to breeding lines (48%, 0.85) and cultivars (50%, 0.81). Application of Bayesian methods for clustering resulted in the robust identification of 20 clusters of accessions, which were unstratified according to origin or user type. This indicates an overlap in genetic diversity despite disruptive selection for fiber versus oil types. Nevertheless, eight clusters contained high proportions (70–100%) of commercial cultivars, whereas two clusters were rich (60%) in landraces. These findings provide a basis for better flax germplasm management, core collection establishment, and exploration of diversity in breeding, as well as for exploration of the role of retrotransposons in flax genome dynamics.




Similar content being viewed by others
References
Allaby RG, Peterson GW, Merriwether DA, Fu YB (2005) Evidence of the domestication history of flax (Linum usitatissimum L.) from genetic diversity of the sad2 locus. Theor Appl Genet 112:58–65
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 17:3389–3402
Antonius-Klemola K, Kalendar R, Schulman AH (2006) TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet 112:999–1008
Belyayev A, Kalendar R, Brodsky L, Nevo E, Schulman AH, Raskina O (2010) Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mob DNA 1:6
Brown AHD, Spillane C (1999) Implementing core collections—principles, procedures, progress, problems and promise. In: Johnson RC, Hodgkin T (eds) Core collections for today and tomorrow. IPGRI, Rome
Chen Y, Schneeberger RG, Cullis CA (2005) A site-specific insertion sequence in flax genotrophs induced by environment. New Phytol 167:171–180
Chen Y, Lowenfeld R, Cullis CA (2009) An environmentally induced adaptive (?) insertion event in flax. Int J Genet Mol Biol 3:38–47
Chennaveeraiah MS, Joshi KK (1983) Karyotypes in cultivated and wild species of Linum. Cytologia 48:833–841
Cieslarová J, Smýkal P, Dočkalová Z, Hanáček P, Procházka S, Hýbl M, Griga M (2010) Molecular evidence of genetic diversity changes in pea (Pisum sativum L.) germplasm after long-term maintenance. Genet Resour Crop Evol. doi:10.1007/s10722-010-9591-3
Cloutier S, Niu Z, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119:53–63
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinform 9:539
Cullis CA (2005) Mechanisms and control of rapid genomic changes in flax. Ann Bot 95:201–206
Diederichsen A (2007) Ex situ collections of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genet Resour Crop Evol 54:661–678
Diederichsen A, Fu YB (2006) Phenotypic and molecular (RAPD) differentiation of four infraspecific groups of cultivated flax (Linum usitatissimum L. subsp. usitatissimum). Genet Resour Crop Evol 53:77–90
Diederichsen A, Hammer K (1995) Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet Resour Crop Evol 42:263–272
Diederichsen A, Raney JP (2008) Pure-lining of flax (Linum usitatissimum L.) genebank accessions for efficiently exploiting and assessing seed character diversity. Euphytica 164:255–273
Durrant A (1962) The environmental induction of heritable changes in Linum. Heredity 17:27–61
Eschholz TW, Stamp P, Peter R, Hund A (2008) Genetic diversity of Swiss maize (Zea mays L. ssp mays) assessed with individuals and bulks on agarose gels. Genet Resour Crop Evol 57:71–84
Evans GM, Durrant A, Rees H (1966) Associated nuclear changes in the induction of flax genotrophs. Nature 212:697–699
Everaert I, de Riek J, de Loose M, van Waes J, van Bockstaele E (2001) Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds 14:69–87
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Frankel OH, Brown AHD (1984) Plant genetic resources today: a critical appraisal. In: Holden JHW, Williams JT (eds) Crop genetic resources: conservation & evaluation. George Allan & Unwin, London, pp 249–257
Fu YB (2002) Redundancy and distinctness in flax germplasm as revealed by RAPD. Plant Genet Resour 4:117–124
Fu YB (2005) Geographic patterns of RAPD variation in cultivated flax. Crop Sci 45:1084–1091
Fu YB, Allaby RG (2010) Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genet Resour Crop Evol 57:667–677
Fu YB, Diederichsen A, Richards KW, Peterson G (2002a) Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPDs. Genet Resour Crop Evol 49:167–174
Fu YB, Peterson G, Diederichsen A, Richards KW (2002b) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Resour Crop Evol 49:253–259
Fu YB, Guerin S, Peterson GW, Diederichsen A, Rowland GG, Richards KW (2003) RAPD analysis of genetic variability of regenerated seeds in the Canadian flax cultivar CDC Normandy. Seed Sci Technol 1:207–211
Fu YB, Legge WG, Clarke JM, Richards KW (2006) Genetic variation and relationships of pedigree-known oat, wheat, and barley cultivars releaved by bulking and single-plant sampling. Genet Resour Crop Evol 53:1153–1164
Gill KS, Yermanos DM (1967) Cytogenetic studies on the genus Linum L. Hybrids among taxa with 15 as the haploid chromosome number. Crop Sci 7:623–627
Goldsbrough PB, Cullis CA (1981) Characterization of the genes for ribosomal RNA in flax. Nucleic Acids Res 9:1301–1309
Grandbastien MA (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3:181–187
Haggans CJ, Hutchins AM, Olson BA, Thomas W, Martini MC, Slavin JL (1999) Effect of flaxseed consumption on urinary estrogen metabolites in postmenopausal women. Nutr Cancer 33:188–195
Hickey M (1988) 100 families of flowering plants, 2nd edn. University Press, Cambridge
Hilakivi-Clarke L, Clarke R, Lippman M (1999) The influence of maternal diet on breast cancer risk among female offspring. Nutrition 15:392–401
Huo H, Conner JA, Ozias-Akins P (2009) Genetic mapping of the apospory-specific genomic region in Pennisetum squamulatum using retrotransposon-based molecular markers. Theor Appl Genet 119:199–212
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Jing R, Vershinin A, Grzebyta J, Shaw P, Smýkal P, Marshall D, Ambrose MJ, Ellis THN, Flavell AJ (2010) The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol Biol 10:44
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc 1:2478–2484
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838
Kalendar R, Lee D, Schulman AH (2009) FastPCR software for PCR primer and probe design and repeat search. Genes Genomes Genomics 3:1–14
Kalendar R, Antonius K, Smýkal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430
Kulpa W, Danert S (1962) Zur Systematik von Linum usitatissimum L. (On the systematics of Linum usitatissimum L.) Kultupflanze. Beiheft 3:341–388
Kvavadze E, Bar-Yosef O, Belfer-Cohen A, Boaretto E, Jakeli N, Matskevich Z, Meshveliani T (2009) 30,000-year-old wild flax fibers. Science 325:1359
Kwon SJ, Hu J, Coyne CJ (2010) Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant genetic resources: characterization and utilization. doi:10.1017/S1479262110000201
Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 2:295–302
Luck JE, Lawrence GJ, Finnegan EJ, Jones DA, Ellis JG (1998) A flax transposon identified in two spontaneous mutant alleles of the L6 rust resistance gene. Plant J 3:365–369
McDill J, Repplinger M, Simpson BB, Kadereit JW (2009) The phylogeny of Linum and Linaceae subfamily Linoideae, with implications for their systematics, biogeography, and evolution of heterostyly. Syst Bot 34:386–405
Menotti-Raymond M, David VA, Pflueger SM, Lindblad-Toh K, Wade CM, O’Brien SJ, Johnson WE (2008) Patterns of molecular genetic variation among cat breeds. Genomics 1:1–11
Muravenko OV, Yurkevich OY, Bolsheva NL et al (2009) Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 135:245–255
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Petit M, Guidat C, Daniel J et al (2010) Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol 186:135–147
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Reif JC, Melchinger AE, Frish M (2005) Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci 45:1–7
Rohlf F (2006) NTSYSpc: numerical taxonomy system (ver. 2.2). Exeter Publishing, Ltd, Setauket
Schneeberger R, Cullis CA (1991) Specific DNA alterations associated with the environmental induction of heritable change in flax. Genetics 128:619–630
Schulman AH, Flavell AJ, Ellis TH (2004) The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol 260:145–173
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10:908–915
Smith JSC, Chin EC, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.) comparisons with data from RFLPS and pedigree. Theor Appl Genet 95:163–173
Smýkal P (2006) Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J Appl Genet 47:221–230
Smýkal P, Hýbl M, Corander J, Jarkovský J, Flavell AJ, Griga M (2008a) Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor Appl Genet 117:413–424
Smýkal P, Horáček J, Dostálová R, Hýbl M (2008b) Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J Appl Genetic 49:155–166
Smýkal P, Kenicer G, Flavell AJ, Kosterin O, Redden RJ, Ford R, Zong X, Coyne CJ, Maxted N, Ambrose MJ, Ellis THN (2011) Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genetic Resources. doi:10.1017/S147926211000033X
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman. San Francisco. SPSS for Windows, Rel. 12.0.1. 2003. SPSS Inc, Chicago
Tanhuanpää P, Kalendar R, Schulman AH, Kiviharju E (2008) The first doubled haploid linkage map for cultivated oat. Genome 51:560–569
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Sci Vol 277:1063–1066
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uysal H, Fu YB, Kurt O, Peterson GW, Diederichsen A, Kusters P (2010) Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57:1109–1119
van Treuren R, van Soest LJM, van Hintum TJL (2001) Marker-assisted rationalisation of genetic resource collections: a case study in flax using AFLPs. Theor Appl Genet 103:144–152
Vavilov NI (1926) Studies on the origin of cultivated plants. Bull Appl Bot 26. USSR, Leningrad
Vromans J (2006) Molecular genetic studies in flax (Linum usitatissimum L.). Ph.D. Thesis, Wageningen University, The Netherlands
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini A (2009) Genetic variability in sunflower (Helianthus annuus L.) and in the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 119:1027–1038
Wiesnerova D, Wiesner I (2004) ISSR-based clustering of cultivated flax germplasm is statistically correlated to thousand seed mass. Mol Biotechnol 26:207–214
Williams IH (1988) The pollination of linseed and flax. Bee World 69:145–152
Wright S (1965) The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution 19:395–420
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Zohary D, Hopf M (2000) Domestication of plants in the Old World: the origin and spread of cultivated plants in West Asia, Europe and the Nile Valley. Oxford University Press, Oxford, p 316
Acknowledgments
This work was financially supported by the Ministry of Education of Czech Republic research project MSMT 2678424601. The technical support of Mrs. L. Vítámvásová and E. Fialová, as well as the laboratory support of Anne-Mari Narvanto and Ursula Lönnqvist of the Institute of Biotechnology, University of Helsinki, are gratefully acknowledged. P.S. acknowledges the OECD fellowship AGR/PR(2006)1-JA00035882, within which flax LTR sequences and IRAP primers were generated in the Schulman laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
Gene Bank accessions
DQ767972, GU735096–GU735098, GU929874–GU929878, GU980587–GU980590
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Smýkal, P., Bačová-Kerteszová, N., Kalendar, R. et al. Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor Appl Genet 122, 1385–1397 (2011). https://doi.org/10.1007/s00122-011-1539-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1539-2