Abstract
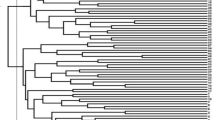
The present investigation aimed to explore the level of genetic diversity, determine the population structure in a larger set of germplasm of linseed using microsatellite marker and identify linked markers through association mapping. A total of 168 accessions of linseed were evaluated for major agro-economic traits and SSRs markers deployed for diversity assessment. A total of 337 alleles were amplified by 50 SSRs ranging from 2 to 13 with an average of 6.74 ± 2.8 alleles per loci. The neighbor joining based clustering grouped all the accessions into three major clusters that were also confirmed by scatter plot of PCoA. While model based clustering determined four sub-populations (K = 4). Further, analysis of molecular variance analysis considering three population showed that maximum variation (79%) was within the population. We identified one putative SSR marker (Lu_3043) linked with days to 50% flowering through both GLM and MLM analysis of association mapping. The results of this preliminary study revealed genetic diversity, population structure in linseed and linked marker which could be utilized in future breeding program.




Similar content being viewed by others

References
Allaby RG, Peterson GW, Merriwether DA, Fu YB (2005) Evidence of the domestication history of flax (Linum usitatissimum L.) from genetic diversity of the sad2 locus. Theor Appl Genet 112:58–65
Allard RW (1999) Principles of plant breeding, 2nd edn. Wiley, New York
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Bickel CL, Gadani S, Lukacs M, Cullis CA (2011) SSR markers developed for genetic mapping in flax (Linum usitatissimum L.). Res Rep Biol 2011:23–29
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cai S, Yu G, Chen X, Huang Y, Jiang X, Zhang G, Jin X (2013) Grain protein content variation and its association analysis in barley. BMC Plant Biol 13:35
Cane MA, Maccaferri M, Nazemi G, Salvi S, Francia R, Colalongo C, Tuberosa R (2014) Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Mol Breed 34:1629–1645
Chandrawati, Maurya R, Singh PK, Ranade SA, Yadav HK (2014) Diversity analysis in Indian genotypes of linseed (Linum usitatissimum L.) using AFLP marker. Gene 549:171–178
Cheng P, William H, Yu M, Coyne CJ, Mazourek M, Grusak MA, Sam F, McGee RJ (2015) Association mapping of agronomic and quality traits in USDA pea single-plant collection. Mol Breed 35:75
Cloutier S, Niu Z, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119:53–63
Cloutier S, Ragupathy R, Niu Z, Duguid S (2010) SSR- based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol Breed 28:437–451
Cloutier S, Ragupathy R, Miranda E, Radovanovic N, Reimer E, Walichnowski A, Ward K, Rowland G, Duguid S, Banik M (2012a) Integrated consensus genetic and physical maps of flax (Linum usitatissimum L.). Theor Appl Genet 125:1783–1795
Cloutier S, Miranda E, Ward K, Radovanovic N, Reimer E, Walichnowski A, Datla R, Rowland G, Duguid S, Ragupathy R (2012b) Simple sequence repeat marker development from bacterial artificial chromosome end sequences and expressed sequence tags of flax (Linum usitatissimum L.). Theor Appl Genet 125:685–694
Cui D, Xu CY, Tang CF, Yang CG, Yu TQ, Xin-xiang A, Cao GL, Xu FR, Zhang JG, Han LZ (2013) Genetic structure and association mapping of cold tolerance in improved japonica rice germplasm at the booting stage. Euphytica 193:369–382
Deng X, Long SH, He DF, Li X, Wang YF, Liu J, Chen XB (2010) Development and characterization of polymorphic microsatellite markers in Linum usitatissmum. J Plant Res 123:119–123
Deng X, Long S, He D, Li X, Wang Y, Hao D, Qiu C, Chen X (2011) Isolation and characterization of polymorphic microsatellite markers from flax (Linum usitatissimum L.). Afr J Biotechnol 10:734–739
Diederichsen A, Ulrich A (2009) Variability in stem fibre content and its association with other characteristics in 1177flax (Linum usitatissimum L.) genebank accessions. Ind Crop Prod 30:33–39
Earl DA, Von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evano method. Conserv Genet Resour 4:359–366
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298
Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman M, Buckler E (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44:1054–1064
Font I Forcada C, Velasco L, Company IRS, Martí IÁF (2015) Association mapping for kernel phytosterol content in almond. Front Plant Sci 6:530
Green A, Chen Y, Singh S, Dribnenki P (2008) Flax. In: Kole C, Hall TC (eds) A compendium of transgenic crop plants. Blackwell, Oxford, pp 199–206
Hickey M (1988) 100 families of flowering plants, 2nd edn. University Press, Cambridge
Jin L, Lu Y, Xiao P, Sun M, Corke H (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121:475–487
Johnson HW, Robinson HF, Comstock RE (1955) Estimates of genetic and environmental variability in Soybean. Agron J 47:314–318
Kale SM, Pardeshi VC, Kadoo NY, Ghorpade PB, Jana MM, Gupta VS (2012) Development of genomic simple sequence repeat markers for linseed using next-generation sequencing technology. Mol Breed 30:597–606
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Newell MA, Cook D, Tinker NA, Jannink JL (2010) Population structure and linkage disequilibrium in oat (Avena sativa L.): implications for genome-wide association studies. Theor Appl Genet 122:623–632
Oh TJ, Gorman M, Cullis CA (2000) RFLP and RAPD mapping in flax (Linum usitatissimum). Theor Appl Genet 101:590–593
Pali V, Verma SK, Xalxo MS, Saxena RR, Mehta N, Verulkar SB (2014) Identification of microsatellite markers for fingerprinting popular Indian flax (Linum usitatissimum L.) cultivars and their utilization in seed genetic purity assessments. Aust J Crop Sci 8:119–126
Pali V, Mehta N, Verulkar SB, Xalxo MS, Saxena RR (2015) Molecular diversity in Flax (Linum usitatissimum L.) as revealed by DNA based markers. Int J Plant Res 28:157–165
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Enfield Science Publishers, Montpellier, pp 43–76
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155:945–959
Rajwade AV, Arora RS, Kadoo NY, Harsulkar AM, Ghorpade PB, Gupta VS (2010) Relatedness of Indian flax genotypes (Linum usitatissimum L.): an inter-simple sequence repeat (ISSR) primer assay. Mol Biotechnol 45:161–170
Roose-Amsaleg C, Cariou Pham E, Vautrin D, Tavernier R, Solignac M (2006) Polymorphic microsatellite loci in Linum usitatissimum. Mol Ecol Notes 6:796–799
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Soto-Cerda BJ, Carrasco RA, Aravena GA, Urbina HA, Navarro CS (2011) Identifying novel polymorphic microsatellites from cultivated flax (Linum usitatissimum L.) following data mining. Plant Mol Biol Rep 29:753–759
Soto-Cerda BJ, Maureira-Butler I, Munoz G, Rupayan A, Cloutier S (2012) SSR-based population structure, molecular diversity and linkage disequilibrium analysis of a collection of flax (Linum usitatissimum L.) varying for mucilage seed-coat content. Mol Breed 30:875–888
Soto-Cerda BJ, Diederichsen A, Ragupathy R, Cloutier S (2013) Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol 13:78
Soto-Cerda BJ, Duguid S, Booker H, Rowland G, Diederichsen A, Cloutier S (2014) Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection. Theor Appl Genet 127:881–896
Spielmeyer W, Green AG, Bittisnich D, Mendham N, Lagudah ES (1998) Identification of quantitative trait loci contributing to Fusarium wilt resistance on an AFLP linkage map of flax (Linum usitatissimum). Theor Appl Genet 97:633–664
Vavilov N (1926) Studies on the origin of cultivated plants, vol 16. Bull Appl Bot Plant Breed, Leningrad, USSR, pp 139-248
Westcott NA, Muir AD (2003) Flax seed lignan in disease prevention and health promotion. Phytochem Rev 2:401–417
Yan WG, Li Y, Agrama HA, Luo D, Gao F, Lu X, Ren G (2009) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol Breed 24:277–292
Yu JM, Pressoir G, Briggs WH, Bi IV, Yamasaki M (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zeist WV, Bakker-Heeres JAH (1975) Evidence for linseed cultivation before 6000 BC. J Archeol Sci 2(3):215–219
Zohary D, Hopf M (2000) Domestication of plants in the old world: the origin and spread of cultivated plants in West Asia, Europe and the Nile Valley. Oxford University Press, Oxford, p 316
Acknowledgements
Authors thank the Director, CSIR-NBRI, Lucknow for providing the facilities to carry out the present investigation. Financial support in form of DST-INSPIRE Fellowship to Chandrawati is gratefully acknowledged.
Author’s contribution
HKY, SAR, RK and SK conceived and designed the research. C, NS, carried out the experiments. PKS provided genetic materials. HKY, VKY, SAR and RK analyzed data. C, NS, SK and HKY wrote the manuscript. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chandrawati, Singh, N., Kumar, R. et al. Genetic diversity, population structure and association analysis in linseed (Linum usitatissimum L.). Physiol Mol Biol Plants 23, 207–219 (2017). https://doi.org/10.1007/s12298-016-0408-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-016-0408-5