Abstract
Our understanding of the evolution of plant sex chromosomes is increasing rapidly due to high-throughput sequencing data and phylogenetic and molecular-cytogenetic approaches that make it possible to infer the evolutionary direction and steps leading from homomorphic to heteromorphic sex chromosomes. Here, we focus on four species of Coccinia, a genus of 25 dioecious species, including Coccinia grandis, the species with the largest known plant Y chromosome. Based on a phylogeny for the genus, we selected three species close to C. grandis to test the distribution of eight repetitive elements including two satellites, and several plastid and mitochondrial probes, that we had previously found to have distinct accumulation patterns in the C. grandis genome. Additionally, we determined C-values and performed immunostaining experiments with (peri-)centromere-specific antibodies on two species (for comparison with C. grandis). In spite of no microscopic chromosomal heteromorphism, single pairs of chromosomes in male cells of all three species accumulate some of the very same repeats that are enriched on the C. grandis Y chromosome, pointing to either old (previous) sex chromosomes or incipient (newly arising) ones, that is, to sex chromosome turnover. A 144-bp centromeric satellite repeat (CgCent) that characterizes all C. grandis chromosomes except the Y is highly abundant in all centromeric regions of the other species, indicating that the centromeric sequence of the Y chromosome diverged very recently.



Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- FISH:
-
Fluorescent in situ hybridization
- PCR:
-
Polymerase chain reaction
- rDNA:
-
Ribosomal DNA
- SDR:
-
Sex-determining region
References
Aliyeva-Schnorr L, Ma L, Houben A (2015) A fast air-dry dropping chromosome preparation method suitable for FISH in plants. JoVE 106:e53470
Bock R, Timmis JN (2008) Reconstructing evolution: gene transfer from plastids to the nucleus. BioEssays 30:556–566
Borchert T, Fuchs J, Winkelmann T, Hohe A (2007) Variable DNA content of Cyclamen persicum regenerated via somatic embryogenesis: rethinking the concept of long-term callus and suspension cultures. Plant Cell Tissue Organ Cult 90:255–263
Cermak T, Kubat Z, Hobza R, Koblizkova A, Widmer A, Macas J, Vyskot B, Kejnovsky E (2008) Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosom Res 16:961–976
Charlesworth D (2016) Plant sex chromosomes. Annu Rev Plant Biol 67:397–420
Chung KS, Hipp AL, Roalson EH (2012) Chromosome number evolves independently of genome size in a clade with nonlocalized centromeres (Carex: Cyperaceae). Evolution 66:2708–2722
Demidov D, Schubert V, Kumke K, Weiss O, Karimi-Ashtiyani R, Buttlar J, Heckmann S, Wanner G, Dong Q, Han F, Houben A (2014) Anti-phosphorylated histone H2AThr120: a universal microscopic marker for centromeric chromatin of mono- and holocentric plant species. Cytogenet Genome Res 143:150–156
Deng CL, Qin RY, Wang NN, Cao Y, Gao J, Gao WJ, Lu LD (2012) Karyotype of asparagus by physical mapping of 45S and 5S rDNA by FISH. J Genet 91:209–212
Fechter I, Hausmann L, Daum M, Sörensen TR, Viehöver P, Weisshaar B, Töpfer R (2012) Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol Gen Genomics 287:247–259
Fraser LG, Tsang GK, Datson PM, De Silva HN, Harvey CF, Gill GP, Crowhurst RN, McNeilage NA (2009) A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics 10:102
Fuchs J, Brandes A, Schubert I (1995) Telomere sequence localization and karyotype evolution in higher plants. P1 Syst Evol 196:227–241
Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nuñez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, Cronk QC (2015) Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol 24:3243–3256
Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jina W (2009) Centromere repositioning in cucurbit species: Implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci USA 106:14937–14941
Harkess A, Mercati F, Shan HY, Sunseri F, Falavigna A, Leebens-Mack J (2015) Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis). New Phytol 207:883–892
Holstein N (2015) Monograph of Coccinia (Cucurbitaceae). PhytoKeys 54:1–166
Holstein N, Renner SS (2011) A dated phylogeny and collection records reveal repeated biome shifts in the African genus Coccinia (Cucurbitaceae). BMC Evol Biol 11:28
Houben A, Wako T, Furushima-Shimogawara R, Presting G, Künzel G, Schubert I, Fukui K (1999) The cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J 18:675–679
Iovene M, Yu Q, Ming R, Jiang J (2015) Evidence for emergence of sex-determining gene(s) in a centromeric region in Vasconcellea parviflora. Genetics 199:413–421
Kafkas S, Khodaeiaminjan M, Güney M, Kafkas E (2015) Identification of sex-linked SNP markers using RAD sequencing suggests ZW/ZZ sex determination in Pistacia vera L. BMC Genomics 16:98
Kejnovsky E, Kubat Z, Hobza R, Lengerova M, Sato S, Tabata S, Fukui K, Matsunaga S, Vyskot B (2006) Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica 128:167–175
Kersten B, Pakull B, Groppe K, Lueneburg J, Fladung M (2014) The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biol 16:411–418
Lappin FM, Medert CM, Hawkins KK (2015) A polymorphic pseudoautosomal boundary in the Carica papaya sex chromosomes. Mol Gen Genomics 290:1511–1522
Marais GA, Forrest A, Kamau E (2011) Multiple nuclear gene phylogenetic analysis of the evolution of dioecy and sex chromosomes in the genus Silene. PLoS One 6:e21915
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Ann Rev Plant Biol 62:485–514
Mrackova M, Nicolas M, Hobza R (2008) Independent origin of sex chromosomes in two species of the genus Silene. Genetics 79:1129–1133
Nagaki K, Walling J, Hirsch C, Jiang J, Murata M (2009) Structure and evolution of plant centromeres. In: Ugarkovic D (ed) Centromere. Structure and evolution. Progress in molecular and subcellular biology, vol 48. Springer, Berlin, pp 154–179
Oyama RK, Clauss MJ, Formanová N (2008) The shrunken genome of Arabidopsis thaliana. Plant Syst Evol 273:257–271
Pellicer J, Kelly LJ, Leitch IJ, Zomlefer WB, Fay MF (2014) A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol 201:1484–1497
Picq S, Santoni S, Lacombe T, Latreille M, Weber A, Ardisson M, Ivorra S, Maghradze D, Arroyo-Garcia R, Chatelet P, This P, Terral JF, Bacilieri R (2014) A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol 14:229
Piednoel M, Aberer AJ, Schneeweiss GM, Macas J, Novak P, Gundlach H, Temsch EM, Renner SS (2012) Next-generation sequencing reveals the impact of repetitive DNA in phylogenetically closely related genomes of Orobanchaceae. Mol Biol Evol 29:3601–3611
Rockinger A, Sousa A, Carvalho FA, Renner SS (2016) Chromosome number reduction in the sister clade of Carica papaya with concomitant genome size doubling. Amer J Bot 103:1–7
Slancarova V, Zdanska J, Janousek B, Talianova M, Zschach C, Zluvova J, Siroky J, Kovacova V, Blavet H, Danihelka J, Oxelman B, Widmer A, Vyskot B (2013) Evolution of sex determination systems with heterogametic males and females in Silene. Evolution 67:3669–3677
Sousa A, Renner SS (2015) Interstitial telomere-like repeats in the monocot family Araceae. Bot J Linn Soc 177:15–26
Sousa A, Fuchs J, Renner SS (2013) Molecular cytogenetics (FISH, GISH) of Coccinia grandis: a ca. 3 myr-old species of Cucurbitaceae with the largest Y/autosome divergence in flowering plants. Cytogenet Genome Res 139:107–118
Sousa A, Bellot S, Fuchs J, Houben A, Renner SS (2016) Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J 88:387–396
Spigler RB, Lewers KS, Main DS, Ashman T-L (2008) Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101:507–517
Steflova P, Hobza R, Vyskot B, Kejnovsky E (2014) Strong accumulation of chloroplast DNA in the Y chromosomes of Rumex acetosa and Silene latifolia. Cytogenet Genome Res 142:59–65
Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C (2007) Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol Gen Genomics 278:221–234
Tennessen JA, Govindarajulu R, Liston A, Ashman T-L (2016) Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex-determining region. New Phytol 211:1412–1423
Tlaskal J (1980) Combined cycloheximide and 8-hydroxyquinoline pretreatment for study of plant chromosomes. Stain Tech 54:313–319
Ventura M, Archidiacono N, Rocchi M (2001) Centromere emergence in evolution. Gen Res 11:595–599
Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, Navajas-Pérez R, Feltus FA, Lemke C, Tong EJ, Chen C, Wai CM, Singh R, Wang ML, Min XJ, Alam M, Charlesworth D, Moore PH, Jiang J, Paterson AH, Ming R (2012) Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A 109:13710–13715
Yin T, Difazio SP, Gunter LE, Zhang X, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang M, Tuskan GA (2008) Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res 18:422–430
Zhang W, Wang X, Yu Q, Ming R, Jiang J (2008) DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res 18:1938–1943
Zhang Q, Liu C, Liu Y, VanBuren R, Yao X, Zhong C, Huang H (2015) High-density interspecific genetic maps of kiwifruit and the identification of sex-specific markers. DNA Res 22:367–375
Acknowledgements
We thank Martina Silber for help in the lab, Andreas Houben and two anonymous reviewers for comments on the manuscript, and the German Science Foundation for funding (DFG RE-603/19-1).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Hans de Jong.
Electronic supplementary material
Fig. S1.
Distribution of 5S and 45S rDNA and Arabidopsis-type telomere sequences on mitotic metaphase chromosomes of Coccinia hirtella (a–c), C. sessilifolia (d–i), and mitotic metaphase (j) and meiotic metaphase I (l, l) of C. trilobata, using FISH. Arrows point to chromosomes with weak 45S rDNA signals. The sex of each karyotype is indicated in the upper right-hand corner. Bars correspond to 5 μm and apply to all metaphases of a row (JPEG 486 kb)
Fig. S2.
Phylogenetic relationships of the studied Coccinia species modified from Holstein and Renner (2011). Red dots mark nodes with molecular-clock inferred divergence times, while the black dot marks the inferred age of the most recent common ancestor of all 25 living species of Coccinia (JPEG 137 kb)
Fig. S3.
Distribution of repeats in Coccinia grandis (e-h; from Sousa et al. 2016) and in C. sessilifolia, using fluorescence in situ hybridization. Compare Table 2 for probe names; an asterisk stands for Ty3/gypsy/, a § stands for Ty1/copia/. The sex of each karyotype is indicated in the upper right-hand corner. Bars correspond to 5 μm (JPEG 301 kb)
Fig. S4.
Distribution of repeats in Coccinia grandis (d-f; from Sousa et al. 2016) and in C. sessilifolia, using fluorescence in situ hybridization. Compare Table 2 for probe names; an asterisk stands for Ty3/gypsy/. Arrows in A point to two chromosomes enriched with the CL86 element. The sex of each karyotype is indicated in the upper right-hand corner. Bars correspond to 5 μm (JPEG 233 kb)
Fig. S5.
Distribution of a Coccinia grandis centromere repeat, called CgCent (CL1 in Table 2), in C. sessilifolia and C. hirtella, using fluorescence in situ hybridization. The arrow in C. grandis points to the Y chromosome. The sex of each karyotype is indicated in the upper right-hand corner. Bars correspond to 5 μm (JPEG 167 kb)
Fig. S6.
Distribution of plastid and mitochondrial sequences in mitotic metaphase chromosomes of Coccinia sessilifolia (a–c) and C. grandis (d-f; from Sousa et al. 2016). IR stands for inverted repeat and SSC for small single copy region. The sex of each karyotype is indicated in the upper right-hand corner. Bars correspond to 5 μm (JPEG 266 kb)
Fig. S7.
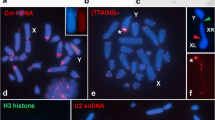
Immunostaining with antibodies against phosphorylated serine 10 of histone H3 (H3Ser10ph), and phosphorylated threonine 120 of histone H2A (H2AThr120ph) on mitotic metaphase chromosomes of Coccinia sessilifolia, C. hirtella, and C. grandis (e-f, from Sousa et al. 2016). Arrows in C. grandis point to its Y chromosome. The sex of each karyotype is indicated in the upper right-hand corner. Bars corresponds to 5 μm (JPEG 272 kb)
Rights and permissions
About this article
Cite this article
Sousa, A., Fuchs, J. & Renner, S.S. Cytogenetic comparison of heteromorphic and homomorphic sex chromosomes in Coccinia (Cucurbitaceae) points to sex chromosome turnover. Chromosome Res 25, 191–200 (2017). https://doi.org/10.1007/s10577-017-9555-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-017-9555-y