Abstract
Key message
We constructed a high-density linkage map comprising 3541 markers developed by specific-locus amplified fragment sequencing, and identified 26 stable QTL including nine novel loci, for fatty acid composition in soybean.
Abstract
Soybean oil quality and stability are mainly determined by the fatty acid composition of the seed. In the present study, we constructed a high-density genetic linkage map using 200 recombinant inbred lines derived from a cross between cultivated soybean varieties Luheidou2 and Nanhuizao, and SNP markers developed by specific-locus amplified fragment sequencing (SLAF-seq). This map comprises 3541 markers on 20 linkage groups and spans a genetic distance of 2534.42 cM, with an average distance of 0.72 cM between adjacent markers. Inclusive composite interval mapping revealed 26 stable QTL for five fatty acids, explaining 0.4–37.0% of the phenotypic variance for individual fatty acids across environments. Of these QTL, nine are novel loci (qLA1, qLNA2_1, qPA4_1, qLA4_1, qPA6_1, qSA12_1, qPA16_1, qOA18_1, and qFA19_1). These stable QTL harbor three fatty acid biosynthesis genes (GmFabG, GmACP, and GmFAD8), and 66 genes encoding lipid-related transcription factors. These stable QTL and tightly linked SNP markers can be used for marker-assisted selection in soybean breeding programs.
Similar content being viewed by others
Introduction
Soybean (Glycine max L. Merr.) is one of the most important oilseed crops worldwide. Providing most of the world’s supply of vegetable protein and oil, soybean accounted for approximately 61% of the world’s oilseed production in 2015 (http://soystats.com). The quality and stability of soybean oil are mainly determined by five predominant fatty acids, viz. palmitic, stearic, oleic, linoleic, and linolenic acids (Lee et al. 2007). Palmitic (16:0) and stearic (18:0) acids are saturated fatty acids, while oleic (18:1), linoleic (18:2), and linolenic (18:3) acids are unsaturated fatty acids. A high proportion of unsaturated fatty acids in the human diet benefits cardiovascular health (Connor 2000; Mensink and Katan 1992). However, polyunsaturated fatty acids, particularly linolenic acid, increase the oxidation of food oils, causing an off-flavor and reducing the shelf life of the oil (Hu et al. 1997; Mounts et al. 1988). Therefore, one important focus of soybean breeding is to improve the fatty acid composition in seed oil.
Fatty acid content is a quantitative trait that depends on the combined effects of several major and minor genes (Bilyeu et al. 2005; Fan et al. 2015; Wang et al. 2014). Therefore, quantitative trait loci (QTL) mapping is an effective method to uncover the genetic basis of fatty acid formation. To date, numerous QTL for fatty acid contents have been detected (Alrefai et al. 1995; Bachlava et al. 2009; Brummer et al. 1997; Diers and Shoemaker 1992; Fan et al. 2015; Hyten et al. 2004; Panthee et al. 2006; Reinprecht et al. 2006; Wang et al. 2012, 2014; Xie et al. 2012). However, these QTL span fairly large genomic regions due to the relatively low density of genetic maps. The relatively low accuracy of QTL mapping using these maps limits not only the identification of fatty acid biosynthesis and regulatory networks, but also the application of these QTL in marker-assisted selection (MAS) breeding efforts in soybean. Recently, putative nucleotide polymorphisms responsible for fatty acid contents, which are usually denoted as quantitative trait nucleotides (QTN), were also identified in genome-wide association study (GWAS) based on population-wide linkage disequilibrium (LD) using soybean natural populations and genome-wide single nucleotide polymorphisms (SNP) (Li et al. 2015). The annotated candidate genes bearing these QTN demonstrated that fatty acid formation is governed by a complex genetic basis in soybean (Li et al. 2015).
With the great development in next-generation sequencing (NGS), several procedures were developed for SNP discovery and genotyping in large population, including restriction-site associated DNA tag sequencing (RAD-seq), genotyping-by-sequencing (GBS), and specific-locus amplified fragment sequencing (SLAF-seq), etc. (Baird et al. 2008; Elshire et al. 2011; Sun et al. 2013). These procedures reduced the genome complexity by digesting genomic DNA with restriction enzymes, and the resultant reduced representation library (RRL) was sequenced to achieve SNP discovery and genotyping in large population. Specifically, a pre-design experiment was performed in SLAF-seq to evaluate restriction enzymes and sizes of restriction fragments using the soybean reference genome sequence, which improved the efficiency of SLAF-seq (Sun et al. 2013). Additionally, the fragments in GBS library were usually selected through PCR amplification (Elshire et al. 2011). In contrast, the fragments in SLAF library were gel-purified, and the fragments with specific size were selected in subsequent sequencing. That will improve the uniformity of fragments in RRL library (Sun et al. 2013). Previously, we developed 200 recombinant inbred lines (RILs) from a cross between two cultivated soybean with different fatty acid compositions. Using 100 of these 200 RILs, We constructed a linkage map consisting of 161 SSR markers, and the QTL for fatty acid composition were identified across 3 years (2009 through 2011) (Fan et al. 2015). In addition, we sequenced 110 of the 200 RILs and the two parents, and developed a high-density genetic map comprising 5785 markers based on the SLAF-seq method (Li et al. 2014).
In the current study, we developed a great number of SNP-based markers using SLAF-seq with an increased mapping population size of 200 RILs to improve the efficiency and accuracy of QTL mapping. The results will benefit the improvement of the fatty acid composition of soybean in breeding project.
Materials and methods
Plant materials and field trials
Two hundred F5:7 RILs developed from the Luheidou2 (LHD2)/Nanhuizao (NHZ) cross together with the parental lines were planted with three replicates in randomized complete blocks at Shunyi Experimental Stations (N40°13′ and E116°34′) in Beijing from 2009 to 2011. Each plot comprised a 2-m row, with 0.5 m apart between rows and a space of 0.1 m between adjacent plants (Fan et al. 2015). Both of the parents are wild type cultivated soybean varieties with black seed coats, but their fatty acid composition differs significantly (Fan et al. 2015).
Fatty acid extraction and determination
The composition of five predominant fatty acids (palmitic, stearic, oleic, linoleic, and linolenic acids) was determined using gas chromatography (Fan et al. 2015). Briefly, 20 g of soybean seeds of each line were ground to a fine powder with a Sample Preparation Mill (Retsch ZM100, Φ = 1.0 mm, Rheinische, Germany). Three hundred milligrams of each powdered sample was transferred to a 2-ml centrifuge tube preloaded with 1.5 ml n-hexane. After vigorous mixing, the mixture was stored at 4 °C for 12 h. Then the samples were centrifuged at 5000×g (room temperature) for 10 min. The supernatant was collected and sodium methoxide solution was added. The mixture was shaken for 1 h on a twist mixer (TM-300, ASONE, Japan) for full methyl esterification of the fatty acids, and centrifuged again at 5000×g for 10 min. The supernatant was collected to determine the composition of the five fatty acids (Fan et al. 2015).
Fatty acid composition was determined using an RTX-Wax Column (30 m × 0.25 mm × 0.25 mm) of gas chromatography (GC-2010, SHIMADZU, Japan). The injection volume was 1 μL. Nitrogen, hydrogen and air were used as carrier gases. The temperature was initially set at 180 °C for 1.5 min, increased to 210 °C at a rate of 10 °C min−1, and maintained at 210 °C for 2 min, increased to 220 °C at a rate of 5 °C min−1 and maintained at 220 °C for 5 min. The area normalization method was used to calculate the composition (percentage of total fatty acids by mass) of the five fatty acids using a GC2010 workstation (Fan et al. 2015).
Specific-locus amplified fragments (SLAF) library construction and sequencing
We previously constructed and sequenced a SLAF library for 110 of the 200 RILs (Li et al. 2014). In our current study, the SLAF library of the remaining 90 RILs and parents was constructed and sequenced, following the same method, with minor modifications: first, the genomic DNA of each sample was digested with a single restriction enzyme, MseI, rather than both EcoRI and MseI according to the pre-design experiment results based on the latest version of the soybean reference genome sequence (Wm82.a2.v1, https://phytozome.jgi.doe.gov) (Schmutz et al. 2010); second, amplified fragments that were 374–474 bp in length instead of 500–550 bp were gel-purified and diluted for pair-end sequencing using an Illumina high-throughput sequencing platform (Illumina, Inc; San Diego, CA, USA).
SLAF-seq data grouping and genotyping
The SLAF-seq data grouping and genotyping of the 90 RILs were performed following the previously reported method (Li et al. 2014; Sun et al. 2013). Briefly, low-quality reads (quality score <20e) were filtered out and then raw reads were sorted to each progeny according to duplex barcode sequences using SLAF_Poly.pl software (Biomarker, Beijing, China). After the barcodes and the terminal 5-bp positions were trimmed from each high-quality reads, clean reads from the same sample were mapped onto the soybean reference genome sequence (Wm82.a2.v1) using SOAP software (Li et al. 2008b; Schmutz et al. 2010). Sequences mapping to the same position with over 95% identity were defined as one SLAF locus. Since soybean is a diploid species and one locus can only contain at most four SLAF tags, groups containing more than four tags were filtered out as repetitive SLAFs, and the SLAFs with 2–4 tags were identified as polymorphic SLAFs (Sun et al. 2013).
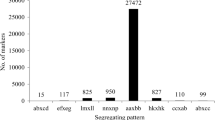
Genotype scoring was then performed using a Bayesian approach to further ensure the genotyping quality (Sun et al. 2013). First, a posteriori conditional probability was calculated using the coverage of each allele and the number of single nucleotide polymorphisms. Then, genotyping quality score translated from the probability was used to select qualified markers for subsequent analysis. Low-quality markers for each marker and each individual were counted and the worse markers or individuals were deleted during the dynamic process. When the average genotype quality scores of all SLAF markers reached the cutoff value, the process stopped. The resultant polymorphic SLAFs were integrated with those of other 110 RILs described in our previous study (Li et al. 2014), and the polymorphic SLAF markers for 200 RILs were obtained. Finally, high-quality SLAF markers for the genetic mapping were filtered by the following criteria: First, average sequence depths should >2-fold in each progeny and >10-fold in the parents. Second, markers with more than 25% missing data were filtered. Third, the Chi-square test was performed to examine the segregation distortion. Markers with significant segregation distortion (P < 0.05) were excluded. The final SNP-based polymorphic SLAF markers were used to construct a high-density linkage map.
Construction of a high-density genetic map
Based on the genotyping data of 200 RILs, a high-density genetic map comprising 20 linkage groups (LGs) was constructed using the Kosambi mapping function of the Joinmap v4.0 software with a LOD threshold of 5.0. The collinearity of 20 LGs with the soybean reference genome was analyzed by plotting the genetic positions of SLAF markers against their physical positions in the soybean reference genome (Wm82.a2.v1).
QTL mapping for fatty acid composition
The additive QTL for palmitic, stearic, oleic, linoleic, and linolenic acids were detected using inclusive composite interval mapping (ICIM) in the BIP (bi-parental populations) model of QTL IciMapping software v4.0 (Li et al. 2008a), with the P values for entering variables (PIN) = 0.05. The threshold of the logarithm of the odds (LOD) scores for evaluating the statistical significance of QTL effects was determined using 1000 permutations at the significance level of 0.05. As a result, a LOD score of 3.3 was used as the threshold to declare the presence of a QTL. As the fatty acid composition is affected by the environments, we focused mainly on the QTL for individual fatty acids identified across multiple environments. The epistatic effects of QTL were identified by the ICIM-EPI method based on the BIP model implemented in QTL IciMapping software v4.0, with PIN = 0.05. The LOD threshold of 5.0 was obtained through 1000 permutation to declare the epistatic QTL at the significance level of 0.05.
Annotation of genes within additive QTL intervals
The sequences within QTL intervals were identified according to the soybean reference genome sequence (Wm82.a2.v1, https://phytozome.jgi.doe.gov), and annotated against Nr (non-redundant), Swiss-Prot, and KOG/COG (clusters of orthologous groups) databases using Blastx program (https://blast.ncbi.nlm.nih.gov).
Results
Phenotypic analysis of fatty acid compositions in soybean RIL population
The fatty acid compositions of 200 RILs were determined from 2009 to 2011. As shown in Table 1, the five predominant fatty acids exhibit broad ranges in 200 RILs. Of them, linoleic acid shows the minimum of coefficient of variance (CV) ranging from 4.3 to 6.9%, while stearic acid presents the maximum of CV ranging from 11.3 to 17.8%. The broad sense heritability of five predominant fatty acids ranged from 0.74 to 0.88 over 3 years, suggesting the fatty acids are mainly controlled by genetic factor (Table 1).
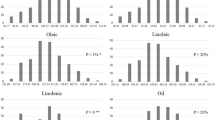
The frequency distributions of the five fatty acids were also analyzed. Almost all fatty acids exhibit continuous and normal distributions from 2009 to 2011, except oleic and linoleic acids in 2011 (Fig. 1; Table 1), suggesting fatty acids are inherited in a quantitative manner. Moreover, significant transgression segregations were also observed in progenies (Fig. 1), suggesting both parents contributed to fatty acid composition.
SLAF-seq and genotyping of soybean RIL population
A total of 281.5 million pair-end reads from SLAF-seq were generated using the Illumina Genome Analyzer IIx in this study. Specifically, 28.4 and 28.9 million reads were generated for female parent LHD2 and male parent NHZ, respectively, while 224.3 million reads were obtained for 90 of the 200 RILs. After filtering, paired-end reads with clear information were mapped to the soybean reference genome (Version of Wm82.a2.v1, https://phytozome.jgi.doe.gov), and 453,524 effective SLAFs were developed. Polymorphisms of the integrated SLAFs were analyzed, and 16,199 polymorphic SLAFs were identified. These polymorphic SLAFs were integrated with the 9948 polymorphic SLAFs identified in other 110 RILs of the population in our previous study (Li et al. 2014), and integrated SLAFs were further screened to filter out markers that were unsuitable for genetic map construction. Finally, 3541 polymorphic SLAFs were obtained to construct a high-density linkage map. All of these markers are of the SNP-type.
Construction of a high-density genetic map in soybean
The genotyping data of the 3541 SLAF markers for 200 RILs were analyzed to determine the order of these SLAF markers in 20 LGs, and a new high-density genetic map was constructed, with a genetic distance of 2534.42 cM (Fig. 2; Supplementary Table S1). The average distance between adjacent markers was 0.72 cM. The largest LG was Gm18, with 339 SLAF markers and a length of 187.47 cM. The smallest LG was Gm04, with 260 SLAF markers and a length of 94.18 cM. The mean chromosome length was 126.72 cM (Table 2). A relatively high collinearity was observed between the 20 LGs and the reference genome (Supplementary Fig. S1), making the annotation of genes within QTL intervals feasible.
High-density genetic map for soybean and 26 stable QTL for fatty acids. The SLAF marker distributions were depicted on the 20 linkage groups (Gm01–Gm20) based on their genetic positions in centiMorgans (cM). The 26 stable QTL for fatty acids, along with their interval distances (cM) are shown between the tightly linked SLAF markers on the right side of each linkage group. The three fatty acid biosynthesis genes are indicated within the QTL intervals in red color by comparing their physical positions with that of SLAF markers in the soybean reference genome (color figure online)
QTL for fatty acid composition
We mapped 316 QTL to 20 soybean chromosomes for the five fatty acids. Of these QTL, 26 were identified for fatty acids across multiple environments, 64 for multiple fatty acids in single environments, and 226 for fatty acids in single environments (Supplementary Table S2). We focused mainly on the 26 stable QTL across multiple environments; these QTL were mapped to all chromosomes except Gm10 and Gm11 (Fig. 2), and the average phenotypic variance explained by individual QTL varied from 0.4 to 37.0% (Table 3). Eight of the 26 QTL explained the high phenotypic variance (>10%) for specific fatty acids. We detected three genes involved in fatty acid biosynthesis, and 66 genes encoding lipid-related transcription factors within the 26 stable QTL intervals (Table 3 and Supplementary Table S3).
Particularly, four stable QTL for palmitic acid (qPA4_1, qPA6_1, qPA8_1, and qPA16_1) explained 1.7–37.0% of the phenotypic variance. qPA8_1 explained an averaged 32.5% of the phenotypic variance for palmitic acid. For stearic acid, four stable QTL (qSA8_1, qSA12_1, qSA14_1 and qSA18_1) accounted for 3.0–28.7% of the phenotypic variance. qSA8_1 and qSA12_1 explained averaged 25.6 and 10.9% of phenotypic variance, respectively, and a 3-oxoacyl-ACP reductase gene (GmFabG, Glyma.12G092900) was found within the genomic region of qSA12_1. For oleic acid, eight stable QTL were detected including qOA7_1, qOA8_1, qOA9_1, qOA13_1, qOA17_1, qOA18_1, qFA19_1, and qFA19_2, explaining 1.3–33.6% of the phenotypic variance. qOA18-1 explained 18.0% of the phenotypic variance on average. An acyl carrier protein gene (GmACP, Glyma.09G060900) was found within the genomic region of qOA9_1. For linoleic acid, six stable QTL were identified, including qFA19_1, FA19_2, qLA1_1, qLA4_1, qLA5_1, and qLA8_1, explaining 0.4–19.3% of the phenotypic variance. qFA19-2 explained an averaged 16.4% of the phenotypic variance. For linolenic acid, six QTL (qLNA2_1, qLNA3_1, qLNA15_1, qLNA16_1, qLNA19_1, and qLNA20_1) explained 2.5–32.3% of the phenotypic variance. qLNA3_1, qLNA15_1 and qLNA19_1 explained on average 12.7, 11.2, and 30.3% of the phenotypic variance, respectively. A ω-fatty acid desaturase gene (GmFAD8, Glyma.03G056700) was found within the genomic region of qLNA3_1 (Table 3). Moreover, qFA19_1 and qFA19_2 contributed to both oleic and linoleic acid composition across multiple environments, suggesting a pleiotropic effect for multiple fatty acids (Table 3).
Epistatic effect on fatty acid composition
Five epistatic QTL were detected for individual fatty acids, explaining 3.4–10.4% of the phenotypic variance (Table 4). Interestingly, a locus at 0 cM on Gm06 had epistatic effects with both loci on Gm07 and Gm10 for stearic acid across two environments (2009 and 2011), indicating a relatively stable epistatic effect (Table 4).
Discussion
SLAF-seq is an effective sequencing-based method for large-scale marker discovery and genotyping. Furthermore, SLAF-seq is a highly efficient approach for marker development that is relatively inexpensive and can be based on large populations (Li et al. 2014; Sun et al. 2013). This method has been widely used to construct high-density genetic maps and identify QTL for agronomic traits and disease resistance in various crops (Han et al. 2015; Qin et al. 2015; Su et al. 2016). However, the application of this method in QTL mapping for soybean fatty acid composition has not been reported. Therefore, we used SLAF-seq to construct a high-density genetic map and identify QTL for fatty acid composition.
The effects of population size and marker density on QTL mapping
Efficiency and accuracy are important for QTL mapping (Li et al. 2010; Stange et al. 2013). We previously performed QTL mapping for fatty acid composition using a genetic linkage map based on 161 SSR marker sets and 100 RILs derived from the LHD2/NHZ cross (Fan et al. 2015). In the current study, all the 200 RILs from the same population were used to construct a new high-density genetic linkage map with 3541 SLAF markers. To analyze the efficiency and accuracy of QTL mapping with different population sizes and marker densities, we made a permutation for QTL detection of fatty acid composition using 100–200 RILs randomly selected from the population and compared the results each other and with our previous study. The number of detected QTL increased significantly with an increase in marker density and population size (Table 5). Therefore, the efficiency of QTL mapping improved significantly with an increase in population size and marker density. Many novel QTL could be identified with the improvement of QTL mapping.
On the other hand, almost all of QTL intervals (45 of the 47 overlapping QTL intervals) were reduced significantly with the increase of marker densities. In fact, 91% of QTL intervals were smaller than 5.0 cM, and 38% of QTL intervals were smaller than 1.0 cM. For instance, qFA8_1 was mapped in an interval of 30.6 cM in our previous study (Fan et al. 2015), whereas the QTL interval was reduced to 1.1 cM in the current study (Supplementary Table S2). The smaller genomic region, in combination with the high collinearity of the genetic map with the reference genome sequence (Supplementary Fig. S1), will facilitate fine mapping of these QTL. That will help uncover the complex networks that govern fatty acid formation and regulation in soybean seeds. Another example, qLNA19_1, explaining an averaged 30.3% of the phenotypic variance for linolenic acid across two environments (2010 and 2011), was identified within a 0.5 cM interval, corresponding to a 55 kb genomic region in the soybean reference genome (Wm82.a2.v1). Therefore, the candidate genes within this region could be identified. Additionally, due to the low density of our previous genetic map, several closely linked QTL were assumed to be a single QTL. When the marker density and population size increased, these QTL could be detected accurately as several individual QTL. For instance, qOA7_2 was mapped in a 17.9 cM interval in our previous study (Fan et al. 2015), whereas two adjacent additive QTL (qFA7_7 and qFA7_8) were detected in the current study (Supplementary Table S2). Therefore, the accuracy of QTL mapping was improved significantly due to the increased population size and marker density.
Comparison analysis revealed the novel stable QTL for fatty acids
According to the SoyBase database (http://soybase.org), hundreds of QTL have been detected for individual fatty acids (Alrefai et al. 1995; Bachlava et al. 2009; Brummer et al. 1997; Diers and Shoemaker 1992; Fan et al. 2015; Hyten et al. 2004; Panthee et al. 2006; Reinprecht et al. 2006; Wang et al. 2012, 2014; Xie et al. 2012). GWAS has also revealed 33 QTN associated with individual fatty acids (Li et al. 2015). We compared the QTL detected in our current study with the QTL and QTN reported previously according to their physical position in the soybean reference genome, and found that 108 of the total 316 QTL are novel (Supplementary Table S2). For the 26 stable QTL detected across multiple environments, nine are novel loci for individual fatty acids. Specifically, for saturated fatty acid, three novel QTL (qPA4_1, qPA6_1, and qPA16_1) were stably identified for palmitic acid, while one novel QTL, qSA12_1, was detected for stearic acid. For unsaturated fatty acid, two novel QTL (qOA18_1 and qFA19_1) were detected for oleic acid; three novel QTL (qFA19_1, qLA1_1 and qLA4_1) were identified for linoleic acid, while one novel QTL, qLNA2_1, contributed to linolenic acid (Table 3). These novel stable QTL could help elucidate the genetic basis of fatty acid accumulation and regulation.
Major stable QTL and tightly linked markers could be applied for marker-assisted selection in soybean breeding programs
We also identified eight major stable QTL explaining the high phenotypic variance (>10%) for fatty acids. Particularly, qPA8_1, qSA8_1, qLNA19_1 explained approximately one-third of the phenotypic variance for palmitic, stearic, and linolenic acids, respectively, across multiple environments. By contrast, our results suggested that the epistatic QTL had less effect on fatty acid contents compared with the additive effects of major QTL (Table 4). Therefore, these major QTL and tightly linked SLAF markers could be applied in MAS in soybean breeding programs.
Gene annotation revealed fatty acid biosynthesis and lipid-related transcription factor genes
By annotating the genes within the 26 QTL intervals aginst Nr, Swiss-Prot, and KOG/COG databases, three essential genes involved in fatty acid biosynthesis (GmACP, GmFabG, and GmFAD8) were identified for oleic, stearic, and linolenic acids within the genomic region of qOA9_1, qSA12_1, and qLNA3_1, respectively. GmACP is an essential substrate that functions in upstream of de novo fatty acid biosynthesis. GmFabG is one of the core enzymes of fatty acid synthase (FAS). It catalyzes the reduction of acetoacetyl-ACP to β-hydroxybutyryl-ACP. In combination with other enzymes of FAS, palmitoyl-ACP (16:0-ACP) is produced (Somerville and Browse 1996). GmFAD8 is a ω-fatty acid desaturase that catalyzes the conversion of linoleic acid (18:2) to linolenic acid (18:3), and thereby is important for linolenic acid accumulation (Somerville and Browse 1996). Consistent with their functions, the QTL harboring these three genes contributed to oleic, stearic, and linolenic acids, respectively. The presence of these genes within the QTL suggests that they may contribute to the major effects of these loci.
In addition to the structural genes involved in fatty acid biosynthesis, 66 genes encoding lipid-related transcription factors, such as MYB, WRKY, bZIP, and bHLH, were also detected within the 26 QTL intervals (Supplementary Table S3). Several transcription factors play essential regulatory roles in fatty acid formation (Baud et al. 2007, 2009; Mendes et al. 2013; Mu et al. 2008; Raffaele et al. 2008; To et al. 2012; Wang et al. 2007). Therefore, the functional validation of these 66 genes in fatty acid regulation will help uncover the complex network underlying fatty acid composition in soybean seed.
In summary, we developed a high-density genetic map comprising 3541 SLAF markers using 200 RILs. With this high-resolution genetic map, we identified 26 stable QTL for fatty acid composition. Nine of these QTL are novel loci for individual fatty acids. Three genes involved in fatty acid biosynthesis (GmACP, GmFabG, and GmFAD8) were found within the genomic region of qOA9_1, qSA12_1, and qLNA3_1, respectively, suggesting they may contribute to the major effect for oleic, stearic, and linolenic acids, respectively. The stable and novel QTL detected in the present study will not only facilitate studies of the genetic basis of fatty acid formation and regulation, but they may also be useful in MAS for the improvement of soybean quality.
Author contribution statement
BL conducted the data analysis, QTL mapping, genomic comparative analysis, and wrote the manuscript. SXF, FKY, and YC extracted the DNA from the RIL populations, performed SNP calling, and developed the genetic linkage map for soybean. SRZ designed and edited the figures. FXH, SRY, and LZW provided advice on experimental design and edited the manuscript. JMS designed, supervised, and financed the work and edited the manuscript. All authors read and approved of the final manuscript.
References
Alrefai R, Berke TG, Rocheford TR (1995) Quantitative trait locus analysis of fatty acid concentrations in maize. Genome 38:894–901
Bachlava E, Dewey RE, Burton JW, Cardinal AJ (2009) Mapping and comparison of quantitative trait loci for oleic acid seed content in two segregating soybean populations. Crop Sci 49:433–444
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376
Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50:825–838
Baud S, Wuilleme S, To A, Rochat C, Lepiniec L (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60:933–947
Bilyeu K, Palavalli L, Sleper D, Beuselinck P (2005) Mutations in soybean microsomal omega-3 fatty acid desaturase genes reduce linolenic acid concentration in soybean seeds. Crop Sci 45:1830–1836
Brummer EC, Graef GL, Orf J, Wilcox JR, Shoemaker RC (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci 37:370–378
Connor WE (2000) Importance of n-3 fatty acids in health and disease. Am J Clin Nutr 71:171S–175S
Diers BW, Shoemaker RC (1992) Restriction fragment length polymorphism analysis of soybean fatty acid content. J Am Oil Chem Soc 69:1242–1247
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Fan S, Li B, Yu F, Han F, Yan S, Wang L, Sun J (2015) Analysis of additive and epistatic quantitative trait loci underlying fatty acid concentrations in soybean seeds across multiple environments. Euphytica 206:689–700
Han Y, Zhao X, Cao G, Wang Y, Li Y, Liu D, Teng W, Zhang Z, Li D, Qiu L, Zheng H, Li W (2015) Genetic characteristics of soybean resistance to HG type 0 and HG type 1.2.3.5.7 of the cyst nematode analyzed by genome-wide association mapping. BMC Genomics 16:598
Hu FB, Stampfer MJ, Manson JE, Rimm E, Colditz GA, Rosner BA, Hennekens CH, Willett WC (1997) Dietary fat intake and the risk of coronary heart disease in women. N Engl J Med 337:1491–1499
Hyten DL, Pantalone VR, Saxton AM, Schmidt ME, Sams CE (2004) Molecular mapping and identification of soybean fatty acid modifier quantitative trait loci. J Am Oil Chem Soc 81:1115–1118
Lee J, Bilyeu KD, Shannon JG (2007) Genetics and breeding for modified fatty acid profile in soybean seed oil. J Crop Sci Biotechnol 10:201–210
Li H, Ribaut J-M, Li Z, Wang J (2008a) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260
Li R, Li Y, Kristiansen K, Wang J (2008b) SOAP: short oligonucleotide alignment program. Bioinformatics 4:713–714
Li H, Hearne S, Banziger M, Li Z, Wang J (2010) Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 105:257–267
Li B, Tian L, Zhang J, Huang L, Han F, Yan S, Wang L, Zheng H, Sun J (2014) Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max. BMC Genomics 15:1086
Li YH, Reif JC, Ma YS, Hong HL, Liu ZX, Chang RZ, Qiu LJ (2015) Targeted association mapping demonstrating the complex molecular genetics of fatty acid formation in soybean. BMC Genomics 16:841
Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating FATTY ACID DESATURASE3. Plant Cell 25:3104–3116
Mensink RP, Katan MB (1992) Effect of dietary fatty acids on serum lipids and lipoproteins. A meta-analysis of 27 trials. Arterioscler Thromb Vasc Biol 12:911–919
Mounts TL, Warner K, List GR, Kleiman R, Fehr WR, Hammond EG, Wilcox JR (1988) Effect of altered fatty acid composition on soybean oil stability. J Am Oil Chem Soc 65:624–628
Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, Zuo J (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148:1042–1054
Panthee DR, Pantalone VR, Saxton AM (2006) Modifier QTL for fatty acid composition in soybean oil. Euphytica 152:67–73
Qin D, Dong J, Xu F, Guo G, Ge S, Xu Q, Xu Y, Li M (2015) Characterization and fine mapping of a novel barley stage green-revertible albino gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics 16:838
Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20:752–767
Reinprecht Y, Poysa VW, Yu K, Rajcan I, Ablett GR, Pauls KP (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome 49:1510–1527
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Somerville C, Browse J (1996) Dissecting desaturation: plants prove advantageous. Trends Cell Biol 6:148–153
Stange M, Utz HF, Schrag TA, Melchinger AE, Wurschum T (2013) High-density genotyping: an overkill for QTL mapping? Lessons learned from a case study in maize and simulations. Theor Appl Genet 126:2563–2574
Su J, Pang C, Wei H, Li L, Liang B, Wang C, Song M, Wang H, Zhao S, Jia X, Mao G, Huang L, Geng D, Fan S, Yu S (2016) Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton. BMC Genomics 17:687
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H, Xu C, Song J, Huang L, Wang C, Shi J, Wang R, Zheng X, Lu C, Wang X, Zheng H (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700
To A, Joubes J, Barthole G, Lecureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24:5007–5023
Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52:716–729
Wang X, Jiang GL, Green M, Scott RA, Hyten DL, Cregan PB (2012) Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol Breed 30:1163–1179
Wang X, Jiang GL, Green M, Scott RA, Hyten DL, Cregan PB (2014) Quantitative trait locus analysis of unsaturated fatty acids in a recombinant inbred population of soybean. Mol Breed 32:281–296
Xie D, Han Y, Zeng Y, Chang W, Teng W, Li W (2012) SSR-and SNP-related QTL underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Mol Breed 30:169–179
Acknowledgements
We would like to thank Professor Xianchun Xia (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for his helpful suggestions regarding the manuscript. This work was supported by the Genetically Modified Organisms Breeding Major Projects (No. 2016ZX08004-003), the National Science and Technology Pillar Program during the Twelfth Five-Year Plan Period of China (2014BAD11B01-x02), Ministry of Science and Technology (2016YFD0100504), the National Nature Science Foundation (No. 31671716, No. 31171576 and No. 31301345), and the Chinese Academy of Agricultural Sciences (CAAS) Innovation Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in regard to this manuscript.
Ethical standards
We declare that these experiments comply with the ethical standards in China.
Additional information
Communicated by Istvan Rajcan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, B., Fan, S., Yu, F. et al. High-resolution mapping of QTL for fatty acid composition in soybean using specific-locus amplified fragment sequencing. Theor Appl Genet 130, 1467–1479 (2017). https://doi.org/10.1007/s00122-017-2902-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2902-8