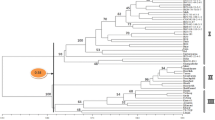
Abstract
Cowpea is an important grain legume and hay crop of many tropical and subtropical regions, especially in the dry savanna region of West Africa. The cowpea gene pool may be narrow because of a genetic bottleneck during domestication. Genetic variation within specific breeding programs may be further restricted due to breeding methods, ‘founder effects’ and limited exchange of germplasm between breeding programs. Genetic relationships among 60 advanced breeding lines from six breeding programs in West Africa and USA, and 27 landrace accessions from Africa, Asia, and South America were examined using amplified fragment length polymorphism (AFLP) markers with six near infrared fluorescence labeled EcoRI + 3/1bases/MseI + 3/1bases primer sets. A total of 382 bands were scored among the accessions with 207 polymorphic bands (54.2%). Despite a diverse origin, the 87 cowpea accessions shared a minimum 86% genetic similarity. Principal coordinates analysis showed clustering of breeding lines by program origin, indicating lack of genetic diversity compared to potential diversity. Accessions from Asia and the Americas overlapped and were distinct from West African breeding lines, indicating that germplasm from Asia and the Americas have common origins outside West Africa. US and Asian breeding programs could increase genetic variability in their programs substantially by incorporating germplasm from West Africa, while national programs in West Africa should consider introgression of Asian germplasm and germplasm from other parts of Africa into their programs to ensure long-term gains from selection.


Similar content being viewed by others
References
Ba FS, Pasquet RS, Gepts P (2004) Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genet Resour Crop Evol 51:539–550
Charcosset A, Moreau L (2004) Use of molecular markers for the development of new cultivars and the evaluation of genetic diversity. Euphytica 137:81–94
Cisse N, Ndiaye M, Thiaw S, Hall AE (1995a) Registration of ‘Mouride’ cowpea. Crop Sci 35:1215–1216
Cisse N, Ndiaye M, Thiaw S, Hall AE (1995b) Registration of ‘Melakh’ cowpea. Crop Sci 37:1978
Coulibaly S, Pasquet RS, Papa R, Gepts P (2002) AFLP analysis of phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor Appl Genet 104:358–366
Ehlers JD, Foster KW (1993) Introgression of agronomic characters from exotic cowpea germplasm into blackeye bean. Field Crops Res 35:43–50
Ehlers JD, Hall AE (1997) Cowpea (Vigna unguiculata L. Walp.). Field Crops Res 53:187–204
Ehlers JD, Hall AE, Patel PN, Roberts PA, Matthews WC (2000) Registration of ‘California Blackeye 27’ Cowpea. Crop Sci 40:854–855
Elawad HOA, Hall AE (2002) Registration of ‘Ein El Gazal’ cowpea. Crop Sci 42:1745–1746
Faris DG (1965) The origin and evolution of the cultivated forms of Vigna sinensis. Can J Genet Cytol 7:433–452
Fatokun CA, Young ND, Myers GO (1997) Molecular markers and genome mapping in cowpea. In: Singh BB, Mohan Raj DR, Dashiell KE, Jackai LEN (eds) Advances in cowpea research, Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS), Ibadan, Nigeria Sayce Publishing, Devon, UK, pp 352–360
Fatokun CA, Danesh D, Young ND, Stewart EL (1993) Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theor Appl Genet 86:97–104
Fotso M, Azanza JL, Pasquet RS, Raymond J (1994) Molecular heterogeneity of cowpea (Vigna unguiculata, Fabaceae) seed storage proteins. Plant Syst Evol 191:39–56
Hall AE, Cisse N, Thiaw S, Elawad HOA, Ehlers JD, Ismail A, Fery R, Roberts P, Kitch LW, Murdock LL, Boukar O, Phillips RD, McWatters KH (2003) Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crops Res 82:103–134
Hooker AL (1973) Southern leaf blight of corn—present status and future prospects. J Environ Qual 1:244–249
Langyintuo AS, Lowenberg-DeBoer J, Faye M, Lambert D, Ibro G, Moussa B, Kergna A, Kushwaha S, Musa S, Ntoukam G (2003) Cowpea supply and demand in West Africa. Field Crops Res 82:215–231
Li C-D, Fatokun CA, Ubi B, Singh BB, Scoles GJ (2001) Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 41:189–197
Ng Q (1995) Cowpea Vigna unguiculata (Leguminosae-Papilionoideae). In: Smartt J, Simmonds NW (eds) Evolution of crop plants, 2nd edn. Longman Scientific and Technical, Singapore, pp 326–332
Ng Q, Padulosi S (1988) Cowpea genepool distribution and crop improvement. In: Ng Q, Perrino P, Attere F, Zedan H (eds) Crop genetic resources of Africa, vol 2, IBPGR, IITA, pp 161–174
Nkongolo KK (2003) Genetic characterization of Malawian cowpea (Vigna unguiculata (L.) Walp.) landraces: diversity and gene flow among accessions. Euphytica 129:219–228
Paczos-Grzeda E (2004) Pedigree, RAPD and simplified AFLP-based assessment of genetic relationships among Avena sativa L. cultivars. Euphytica 138:13–22
Padi FP, Denwar N, Kaleem K, Salifu AB, Clottey B, Kombiok J, Haruna M, Hall AE, Marfo KO (2004a) Registration of ‘Apagbaala’ cowpea. Crop Sci 44:1486
Padi FP, Denwar N, Kaleem K, Salifu AB, Clottey B, Kombiok J, Haruna M, Hall AE, Marfo KO (2004b) Registration of ‘Marfo-Tuya’ cowpea. Crop Sci 44:1486–1487
Padulosi S, Ng Q (1997) Origin, taxonomy, and morphology of Vigna unguiculata L. Walp. In: Singh BB, Mohan Raj DR, Dashiell KE, Jackai LEN (eds) Advances in cowpea research, Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS), Ibadan, Nigeria Sayce Publishing, Devon, UK, pp 1–12
Panella L, Gepts P (1992) Genetic relationship within Vigna unguiculata (L.) Walp. based on isozyme analyses. Genet Resour Crop Evol 39:71–88
Pant KC, Chandel KPS, Joshi BS (1982) Analysis of diversity in Indian cowpea genetic resources. SABRO J 14:103–111
Pasquet RS (1998) Morphological study of cultivated cowpea Vigna unguiculata (L.) Walp. Importance of ovule number and definition of cv gr Melanophthalmus. Agronomie 18:61–70
Pasquet RS (1999) Genetic relationships among subspecies of Vigna unguiculata (L.) Walp. Based on allozyme variation. Theor Appl Genet 98:1104–1119
Pasquet RS (2000) Allozyme diversity of cultivated cowpea Vigna unguiculata (L.) Walp. Theor Appl Genet 101:211–219
Perrino P, Laghetti G, Spagnoletti Zeuli PL, Monti LM (1993) Diversification of cowpea in the Mediterranean and other centers of cultivation. Genet Resour Crop Evol 40:121–132
Purseglove JW (1968) Tropical crops-dicotyledons. Longman Group Ltd, London
Rohlf FJ (2000) NTSYSpc, numerical taxonomy and multivariate analysis system, version 2.1 user guide. Exeter Software, Setauket, New York, USA
Simmonds NW, Smartt J (1999) Principles of crop improvement. Blackwell Science Ltd, Oxford, UK
Singh BB, Ntare B (1985) Development of improved cowpea varieties in Africa. In: Singh SR, Rachie KO (eds) Cowpea research, production and utilization. Wiley, New York, pp 106–115
Singh BB, Ehlers JD, Sharma B, Freire-Filho FR (2002) Recent progress in cowpea breeding. In: Fatokun CA, Tarawali S, Singh BB, Kormawa PM, Tamo M (eds) Challenges and opportunities for enhancing sustainable cowpea production. International Institute of Tropical Agriculture, Ibadan, Nigeria, pp 22–40
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. W.H. Freeman and Company, San Francisco, USA
Steele WM (1976) Cowpea, Vigna unguiculata (Leguminosae-Papillionatae). In: Simmonds NW (eds) Evolution of crop plants. Longman, London
Tarawali SA, Singh BB, Gupta SC, Tabo R, Harris F, Nokoe S, Fernández-Rivera S, Bationo A, Manyong VM, Makinde K, Odion EC (2002) Cowpea as a key factor for a new approach to integrated crop–livestock systems research in the dry savannas of West Africa. In: Fatokun CA, Tarawali S, Singh BB, Kormawa PM, Tamo M (eds) Challenges and opportunities for enhancing sustainable cowpea production. International Institute of Tropical Agriculture, Ibadan, Nigeria, pp 233–251
Tosti N, Negri V (2002) Efficiency of three PCR-based markers in assessing genetic variation among cowpea (Vigna unguiculata subsp. unguiculata) landraces. Genome 45:268–275
Vaillancourt RE, Weeden NF (1992) Chloroplast DNA polymorphism suggests a Nigerian center of domestication for the cowpea, Vigna unguiculata (Leguminosae). Am J Bot 79:1194–1199
Vaillancourt RE, Weeden NF, Barnard JD (1993) Isozyme diversity in the cowpea species complex. Crop Sci 33:606–613
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kupier M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Zeid M, Schön CC, Link W (2003) Genetic diversity in recent elite faba bean lines using AFLP markers. Theor Appl Genet 107:1304–1314
Acknowledgements
The authors thank Drs. N. Cisse, J. Ouedraogo, I. Drabo, B.B. Singh, O. Boukar for supplying cowpea breeding lines used in the present study. This research was supported in part by a grant from Bean/Cowpea Collaborative Research Support Program, USAID Grant no. DAN-G-SS-86-00008-00. The opinions and recommendations are those of the authors and not necessarily those of USAID.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fang, J., Chao, CC.T., Roberts, P.A. et al. Genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genet Resour Crop Evol 54, 1197–1209 (2007). https://doi.org/10.1007/s10722-006-9101-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-006-9101-9