Abstract
Cowpea is a grain legume that is commonly grown in Sub-Saharan Africa (SSA) for a variety of uses including food, feed, and soil fertility enhancement through nitrogen fixation. Genetic improvement in cowpea can be accomplished by creating breeding populations from high-performing but genetically diverse parental lines. The goal of this study was to use single nucleotide polymorphic markers (SNPs) to estimate genetic diversity and population stratification among cowpea parental genotypes from five countries. A total of 2,746 SNP markers were used to analyze 62 cowpea genotypes that differed in key characteristics. Only 55% of the SNP markers were polymorphic across the 62 genotypes and met the selection requirement. The gene diversity varied from 0.52 to 0.57 with an average of 0.55, while the polymorphic information content ranged from 0.46 to 0.51 with an average of 0.49, showing that the cowpea parental lines are genetically diverse. The analysis of molecular variance (AMOVA) found high significant (p < 0.001) variation among genotypes and non-significant variation across genotypes depending on country of origin. The cluster analysis for the 62 genotypes using the unweighted pair group procedure with arithmetic mean approach divided the genotypes into two distinct groups. Genotypes from the same geographical regions tended to cluster together and there was a significant level of genetic variation among genotypes from Uganda and Nigeria. The SNP markers utilized in this study were effective at distinguishing genotypes and may be used to estimate the diversity in the cowpea core collection. The genetic diversity in this collection is important for future development of improved varieties of cowpea in Uganda.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea (Vigna unguiculata (L.) Walp.; 2n = 2x = 22) is a major legume crop produced worldwide under low-input production systems in arid and semi-arid agroecologies. It is an important annual pulse crop farmed in Latin America, South Asia, and Africa’s arid tropical climates (Boukar et al. 2019). The crop is mostly cultivated for its grain, which has high protein (20–32%) and carbohydrate content (50–60%). It is also rich in vitamin C, iron, and zinc found both in the grains and leaves (Gonçalves et al. 2016). According to Jayathilake et al. (2018), cowpea, which is known as “poor man’s meat” in many developing countries, plays a significant role in the human nutrition. The crop is also a key component of traditional cropping systems due to its ability to fix atmospheric nitrogen via symbiosis with rhizobium, which results in soil fertility enhancement, and important in smallholder farming systems where little or no fertilizer is applied (Bado et al. 2006).
Global production of cowpea is projected to reach 12.3 million tons by 2030 (Boukar et al. 2016). It is a versatile crop that provides revenue to millions of smallholder farmers as well as traders who sell healthy grain and leaves as fresh vegetables. Its production and consumption are concentrated in Sub-Saharan Africa, especially West Africa and East Africa, where its nutritional value and drought tolerance traits place it in a unique position in the continent’s attempts to develop nutrition-sensitive food systems (Boukar et al. 2016; Muñoz-Amatriaín et al. 2017). This is especially crucial for lowering malnutrition in pregnant or breastfeeding mothers and children under the age of five (Gomes et al. 2019).
Despite its African origins, the domestication center of cowpea remains unknown (Carvalho et al. 2017); however, it is thought to be in East or West Africa, where there is substantial morphological and genetic diversity is the centre of orgin, followed by a sub-domestication zone in India (Xiong et al. 2016). According to Bado et al. (2006), the selection/domestication of two more subspecies in India, the yard-long bean (Vigna unguiculata subsp. sesquipedalis (L.) Verdc.), and the casting accessions from Europe often cluster with those from West Africa.
Cowpea breeding began in the 1960s, primarily in African nations such as Nigeria, Senegal, Uganda, and Tanzania (Hall 2012). Cowpea breeding has encountered various challenges, including a narrow cowpea gene pool due to a genetic bottleneck during domestication, reduced genetic variability due to “founder effects,” and limited germplasm interchange (Carvalho et al. 2017). Furthermore, the crop has received little attention and has remained an orphan crop in terms of research in a number of countries (Ketema et al. 2020). Efforts have been made in Uganda to enhance cowpea genetic diversity by introducing exotic genetic materials from the International Institute of Tropical Agriculture (IITA), University of California Riverside, and other African national breeding programs, including Ghana, Kenya, and Nigeria, with most of them being breeding lines.
According to Boukar et al. (2019) and Fatokun et al. (2018), the recent advancement in genomics and next-generation sequencing (NGS) technologies have resulted in the generation of high, medium, and low-density molecular markers that are frequently utilized for analyzing genetic diversity in germplasm, genetic fingerprinting, QTL mapping, and gene discovery. In cowpea, several marker technologies have been deployed including random amplified polymorphic DNA (RAPD) (Olufisayo et al. 2016), restriction fragment length polymorphism (RFLP) (Boukar et al. 2016), amplified fragment length polymorphism (AFLP) (Olufisayo et al. 2016; Wamalwa et al. 2016), single sequence repeat (SSR) (Ragul et al. 2018), and single nucleotide polymorphisms (SNPs) (Chidebe et al. 2018).
Among the molecular markers, SNPs have been widely used for analyzing genetic diversity of populations compared to other markers such as AFLP and SSR (Varshney et al. 2007) because of their abundance in the genome of plants and other organisms as well as high throughput (Mammadov et al. 2012). Among the plethora of SNP genotyping platforms, the Diversity Array Technology (DArT-Seq) has been widely used by breeders to generate thousands of SNP markers and to assess the genetic diversity of panels and breeding populations in many crops including maize (Ayesiga et al. 2022), yams (Agre et al. 2019; Bhattacharjee et al. 2020; Amponsah et al. 2023), and cassava (Xia et al. 2005; Adu et al. 2021). DArT-Seq is a hybridization-based DNA sequencing technology that is high-throughput, highly reproducible, at low-cost. In addition, DArT-Seq does not require prior sequence information for detecting SNPs associated with loci/alleles for traits of interest (Nadeem et al. 2018).
The objective of this study was to determine the level of genetic diversity and population structure of cowpea parental lines assembled for breeding in Uganda. Assessing genetic diversity within cowpea germplasm assembled from different geographical locations will help in the selection of a core cowpea germplasm for breeding purposes. This will help guide future development of improved cowpea varieties for Uganda and the region.
Materials and Methods
Genetic Materials and Leaf Sampling
A total of sixty-two (62) genotypes of Vigna unguiculata (L.) Walp, originating from Uganda (23 genotypes; 37%), Ghana (3 genotypes; 0.05%), Nigeria (31 genotypes; 50%), Kenya (2 genotypes; 0.03%), and the United States (3 genotypes; 0.05%), were sampled (Table 1) from a collection available at the Makerere University’s Makerere Regional Centre for Crop Improvement (MaRCCI). The bulk of the genotypes (77.4%) were breeding lines actively utilized in breeding efforts from their various origins, while 11.3% of the genotypes were landraces.
The 62 genotypes were planted in a completely randomized design at MaRCCI in the screen house. Cowpea leaf samples were collected using a method described by KBS-9370–001 for leaf sample collection for DNA extraction. Using a leaf puncher, leaf samples from three (3) tagged plants were collected onto each of the 96-well plates five weeks after planting. The leaf samples were then oven-dried at 80 °C and sent for genotyping at SEQART AFRICA, based at the International Livestock Research Institute (ILRI), Nairobi.
Cowpea DNA Extraction and DArTseq Genotyping
Cowpea genomic DNA was extracted from each leaf sample using a NucleoMag Plant DNA extraction kit (Mag-Bind® Plant DNA DS 96 Kit). The DNA quality for each leaf sample was quantified using NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA), and visualization on 0.8% agarose gel before the concentrations of genomic DNA was adjusted to 100 ng/µl. DNA library construction was performed using the genomic complexity reduction technology (Kilian et al. 2016), which uses a combination of PstI and MseI enzymes for DNA digestion. Following the ligation of barcoded adapters and common adapters, adapter-ligated fragments were amplified using PCR. The libraries were purified and quantified for cluster creation using an automated clonal amplification system (cBOT Illumina) and then sequenced using the Illumina Hiseq2500, an NGS platform (Kilian et al. 2016). The DArTseq markers were scored using DArTsoft14, which is an in-house marker scoring pipeline (Kilian et al. 2016). SilicoDArT markers and SNP markers were both scored as binary for the presence/absence (1 or 0, respectively) of the restriction fragment with the marker sequence in the genomic version of the sample.
Filtering, Genetic Diversity Analyses, and Determination of Population Stratification
TASSEL (v5.2.52) was used for SNP data quality verification and filtering as explained by Bradbury et al. (2007). SNP markers having more than 20% missing data, a minor allele frequency (MAF) of <0.05, with unknown physical position on the chromosomes across samples (genotypes) were deleted. The k-nearest neighbor genotype imputation approach was used to further impute the SNP data in TASSEL (v5.2.52) (Bradbury et al. 2007). A total of 2,746 SNPs were selected and utilized in subsequent analysis. TASSEL (v5.2.52) was used to analyze SNP marker information and minor allele frequencies, and PowerMarker (v3.25) was used to estimate polymorphism information content (PIC) (Liu and Muse 2005). The “adegenet” package in R was used to calculate observed and expected heterozygosity (R Core Team 2020).
Bayesian model-based clustering of the STRUCTURE program (v2.3.4) was used to estimate the number of hypothetical subpopulations (K) (Evanno et al. 2005; Porras-Hurtado et al. 2013). The STRUCTURE analysis used a burn-in period of 10,000 Markov-chain Monte Carlo iterations using an admixture model based on the Hardy–Weinberg equilibrium and its associated allele frequencies.
Ten population numbers (K ranging from 1 to 10) were evaluated, and each K was separately run ten times. The structure outputs were further analysed using STRUCTURE HARVESTER (Earl and vonHoldt 2012), which allowed the optimum K value to be identified as a distinct peak in the change of likelihood (K). To categorize the sixty-two genotypes in the different population groups given by STRUCTURE HARVESTER, a membership threshold (qi) of 0.6 was used, and genotypes that fell below the threshold for all groups were deemed admixture genetic material.
In addition, to complement the STRUCTURE analysis, a discriminant analysis of principal components (DAPC) was performed in R using the “adegenet” package (R Core Team 2020) to determine the optimal number of clusters inferred using the K-means analysis by varying the possible number of clusters from 1 to 10. DAPC scatter plots were prepared for the clusters identified by K-means using the first ten main components.
Furthermore, GenAlEx 6.503 (Peakall and Smouse 2012) was used to perform an analysis of molecular variance (AMOVA) (Reyes-Valdés et al. 2013) to examine population divergence among the genetic groupings found by the cluster analysis. The marker datasets were numerically coded as A = 1, C = 2, G = 3, and T = 4 before AMOVA (Blyton and Flanagan 2006) was performed. AMOVA with 999 permutations was used with the numerically coded marker data. The genetic differences were divided into two categories: variance across populations (PhiPR) and variation within populations (PhiPT). Finally, phylogenetic connections among genotypes were constructed in PowerMarker (v3.25) software using a Euclidean distance matrix (Liu and Muse 2005). Molecular Evolutionary Genetics Analysis (MEGA-X v10.1.8) software was used to show the generated tree (Kumar et al. 2018).
Results
Quality, Diversity, and Functional Characterization of DArTseq-SNPs on Cowpea Chromosome
The 62 cowpea genotypes yielded 7,304 unfiltered SNPs, of which 2,746 good-quality SNPs (37.5%) spread over the 11 chromosomes of Vigna unguiculata were retained for further analysis after filtering (Fig. 1 and Table 2). Chromosome 3 had the highest number of SNPs (338 SNPs; 12.3%), followed by chromosome 4 (332 SNPs; 12.2%) and chromosome 7 (331 SNPs; 12.1%), while chromosome 2 (164 SNPs; 0.06%) had the fewest SNPs (Fig. 1 and Table 2). The polymorphism information content (PIC) as a measure SNP quality indicator varied from 0.47 to 0.51 with a mean value of 0.49. The expected heterozygosity ranged from 0.48 to 0.56, with a mean value of 0.53. On the other hand, the observed heterozygosity varied from 0.04 to 0.07, with a mean of 0.06. Likewise, the average minor allele frequency (MAF) was 0.25 and ranged from 0.21 to 0.29 (Table 2).
Population Differentiation and Genetic Relation Between Cowpea Genotypes
The analysis of molecular variance (AMOVA) found that 97% of the variability was within each genetic group and just 7% was among the five populations/genetic groupings based on geographic origin. Cluster analysis was used to classify the 62 cowpea genotypes into two major clusters (clusters A and B) with sub-clusters within the main clusters (Fig. 2). According to the clustering approach utilized, only one genotype (AC20C042) originated from Nigeria, whereas all six genotypes in cluster A were derived from Uganda. The largest cluster, however, was cluster B, which included genotypes from all geographical regions represented. Cluster B was divided into two subclusters, with one Nigerian genotype standing out (Fig. 2). There was no link between the origin of the cowpea genotypes and the cluster grouping (Table 3).
Population Structure and Discriminate Analysis of Principal Coordinate of Cowpea Genotypes
The STRUCTURE analysis calculation of the populations at each K-value and the membership coefficients (qi) was highly informative. Simulations (logarithm probability relative to standard deviation, K) computed using SNP markers revealed a strong peak at K = 3 and K = 6, which indicated the optimal number of sub-populations (K = 6).
At K = 3, POP I, POP II, and POP III (S1 Table) had 37 (59.7%), 11 (17.7%), and 14 (22.6%) genotypes, respectively. Most genotypes detected in POP I were from Uganda (26 genotypes; 70.2%), followed by Nigeria (8 genotypes; 21.6%) and Kenya (one genotype; 0.27%) (Fig. 3B and Table S1). Most of the genotypes discovered in POP II (7 genotypes; 63.6%) originated from Nigeria, with the remaining sites each having only one genotype. Similarly, genotypes derived from Nigeria dominated POP III (12 genotypes; 85.7%), with the remaining sources represented by only one genotype each.
At ΔK = 6, all the genotypes in POP I (5 genotypes; 0.8%) originated from Nigeria and were the least. POP II had the most genotypes in K = 6, with 16 genotypes (25.8%). Nigeria (8 genotypes) and Uganda (7 genotypes) had the most genotypes in this population, with Kenya having only one genotype. This population was followed by POP IV, which had a total of 15 genotypes (24.2%), the majority of which came from Nigeria (7 genotypes) and Uganda (7 genotypes). POP III and POP VI each had 9 genotypes (14.5%), and the bulk was from Nigeria, with Ghana and Kenya each having only one genotype (Fig. 3B and Table S1). Within the highest peak delta at 6, the expected heterozygosity ranged from 0.06 to 0.38 for POP II and POP III (Table S1), respectively. The measure of population differentiation due to genetic structure (Fst) from the studied population was least observed for POP III (Fst = 0.01) and the highest was in POP II (Fst = 0.96) (Table 4).
Discriminant Analysis Principal Component (DAPC)
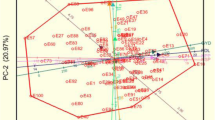
The DAPC method was further conducted to determine the sub-clusters at K = 2 (Fig. 4A), K = 3 (Fig. 4B), K = 4 (Fig. 4C), and K = 5 (Fig. 4D). The summary of the DAPC cluster groupings and probabilities of cluster membership allocations of cowpea genotypes at K = 2, 3, 4, and 5 are presented in the Table S2. Based on the possibility of cluster membership assignment, DAPC cluster grouping both K = 3 and K = 5 represented a good fit.
Discriminant analysis of principal components (DAPC). A Discriminant analysis of principal components with K = 2. B Discriminant analysis of principal components with K = 3. C Discriminant analysis of principal components with K = 4. D Discriminant analysis of principal components with K = 5. The axes represent the first two linear discriminants. Each color represents a cluster and each does represent an individual
The DAPC biplot, together with the plot of individual densities on the first discriminant function, demonstrated an unambiguous division of the cowpea genotypes into three clusters with minimum admixed individuals at K = 3. At K = 3, cluster one had 37 genotypes (59.7%), including 11 genotypes (29.7%) from Nigeria and the bulk (21 genotypes; 56.8%) from Uganda, followed by Ghana (2 genotypes; 5.4%), Kenya (2 genotypes; 5.4%), and the United States (one genotype, 2.7%) (Fig. 4B). Cluster two included just 8 genotypes (12.9%). However, in cluster three, 17 genotypes (27.4%) were found, the bulk of which were from Nigeria (14 genotypes). The results of the DAPC analysis agreed with the conclusions of the STRUCTURE analysis.
Furthermore, the five discriminant functions (Fig. 4D) produced at K = 5 that described cluster one (29 genotypes; 46.8%), cluster two (4 genotypes; 6.5%), cluster three (8 genotypes; 12.9%), cluster four (8 genotypes; 12.9%), and cluster five (13 genotypes; 20.9%) were highly informative. Cluster one which contributed the highest number of genotypes (29 genotypes; 46.8%) consisted of primarily cowpea genotypes sourced from Uganda (12 genotypes; 41.4%) and Nigeria (12 genotypes; 41.4%) with only 2 genotypes (7.3%) sourced from Ghana and Kenya, respectively. However, the cluster groupings and probabilities of cluster membership allocations of genotypes in K = 5 were maintained and equally represented in K = 4 with only cluster three with Nigeria genotypes (Fig. 4D).
Discussion
Genetic diversity is a critical aspect of plant breeding as it provides a pool of variation that can be used to develop improved cultivars with desirable traits (Grzebelus et al. 2014). In the case of cowpea, which is a staple crop in many African countries, understanding the genetic diversity of parental lines is crucial for developing high-yielding and disease-resistant cultivars (Seo et al. 2020). Information on germplasm diversity is key to guiding effective and long-term breeding strategies. In this study, we examined genetic diversity among core parental cowpea genotypes, using informative molecular markers as a critical step toward management, genetic improvement, and conservation of cowpea germplasm in Uganda.
Over 2,000 filtered SNPs were used in this study to determine the degree and distribution of genetic diversity among cowpea genotypes assembled from five different geographical sources. Of the total markers genotyped, only 55% were observed to be polymorphic (MAF ≥ 2%). However, 76% and 63% polymorphism have been reported using inter simple sequence repeats (ISSR) markers within cowpea collections from Nigeria and Brazil (Dias et al. 2015) and indigenous cowpea variants from Africa (Ghalmi et al. 2010). ISSR markers’ prominent level of polymorphism may be due to their substantial number of alleles and abundance in the genome (Varshney et al. 2007).
In this study, the observed genetic distance between pairs of parental cowpea genotypes was minimal, ranging from 0.07 to 0.26 with a mean value of 0.21. Fatokun et al. (2018) previously observed low genetic diversity from an investigation of 370 worldwide cowpea samples. Similarly, Wang et al. (2016) discovered that cowpea accessions in the USDA germplasm collection have little genetic diversity and a short genetic distance. Huynh et al. (2013), on the other hand, discovered a wider range of genetic distances ranging from 0.01 to 0.72 based on common alleles among cowpea landraces from 56 countries. According to Muñoz-Amatriaín et al. (2017), greater genetic diversity is identified when there are more accessions from diverse origins; moreover, cowpea is a selfing crop; thus, the low genetic diversity could be attributed to its reproductive biology and not only because they came from narrow background. Only genotypes from five countries, Uganda, Nigeria, Kenya, Ghana, and the USA, were utilized in this investigation, with moderate genetic diversity. The low genetic distance observed between pairs of parental cowpea genotypes can have several implications for Uganda’s cowpea breeding program, including reduced genetic diversity, difficulty in tracking desirable traits, and increased susceptibility to diseases and environmental stress (Talabi et al. 2017). To overcome these challenges, the breeding programs could consider incorporating more diverse sources of genetic variation, such as wild relatives, into the breeding program to increase the genetic diversity of the parental lines.
In this study, the PIC ranged from 0.47 to 0.51, with an average of 0.49. Desalegne et al. (2016) observed high mean PIC values ranging from 0.23 to 0.68 when employing SSR markers to assess genetic diversity in Ethiopian cowpea germplasm. The moderate variability among the parental lines might be attributed to the self-pollinated nature of cowpea or to the species’ very modest effective population size, which could reduce the diversity of genetic variation that is passed down from one generation to the next (Damarany et al. 2018; Ndjiondjop et al. 2017) and this bottleneck could be induced by a single domestication event in this crop (Gbedevi et al. 2021).
The low amount of polymorphism in cowpea breeding efforts can lead to a small pool of variation that can be leveraged to create better cultivars. Finding parents that exhibit the favorable qualities required for a particular breeding aim, such as high yield, disease resistance, or adaptability to certain growing circumstances, may become more difficult as a result (Acquaah 2012). Cowpea breeding strategies may consider including wild relatives or other various sources of variation in the breeding program to optimize the potential for improvement. This would enhance the likelihood of identifying parents with the desired qualities and provide researchers with a larger variety of variations to work with (Boukar et al. 2019). Additionally, the favorable agronomic and horticultural characteristics of the original cultivar may be preserved while the required traits are incorporated into elite lines using backcrossing and recurrent selection processes.
AMOVA analysis revealed a moderate but significant divergence in cowpea accessions from five different geographical locations. Significant variations were seen among populations and individuals, as well as across individuals (p = 0.05). Individual genetic variance (97% of total variation) was significantly larger than population variance (3% of total variation). Fatokun et al. (2018) and Chen et al. (2017) discovered related results, with the greatest variability identified among accessions compared to within and across populations. As a result, individual variation rather than geographical alignment accounts for most of the genetic variation seen in cowpea.
The findings of this study on genetic variations across cowpea genotypes might have significant ramifications for Uganda’s cowpea breeding efforts. High genetic variability across genotypes can provide a huge pool of variation for developing superior cultivars, but low genetic diversity might restrict the possibility of improvement (Asiedu et al. 1998). These findings can help guide breeding decisions and the selection of parental lines for crosses to accomplish certain breeding objectives. The significant variation among individual genotypes, on the other hand, is due to germplasm sharing either among breeding programs or among smallholder farmers across geographic areas. Furthermore, the limited degree of diversity found across areas might be due to strong gene flow within regions, leaving limited opportunity for genetic differentiation along geographical lines like previous findings (Wamalwa et al. 2016). Because of the low levels of divergence across geographic areas and the high levels of variation within regions, Xiong et al. (2016) indicated that a large random sample would capture the majority of the genetic variation among cowpea genotypes in each region. In addition, excessive germplasm exchange can also lead to a loss of genetic diversity, as farmers may adopt a limited range of varieties and abandon other, potentially valuable, varieties (Teeken et al. 2018). This can reduce the pool of genetic resources available for crop improvement and limit the adaptability of crops to changing environmental conditions. The high amount of germplasm exchange among smallholder farmers across geographic areas can have both positive and negative implications for crop improvement and genetic diversity. To maximize the benefits of germplasm exchange while minimizing potential negative impacts, it is important to promote exchange among farmers while also promoting the conservation of local genetic resources. This can help to ensure that farmers have access to a wide range of genetic resources and that crop improvement efforts are sustainable in the long term.
The grouping patterns of 62 cowpea genotypes from various geographic locations indicated the presence of two separate groups. The observed clustering pattern was not entirely compatible with the genotypes’ geographic origins. The accessions were categorized in the current investigation regardless of where they were collected. The proximity of the accessions in the UPGMA tree revealed a similar genetic composition among several of the genotypes. This might be attributed to seed exchange among breeders and the farmer-to-farmer seed trade system, both of which are common in Sub-Saharan Africa.
SNP-based population structure analysis was useful for preserving and monitoring the genetic variety necessary for a strong breeding program (Eltaher et al. 2018). The SNP markers utilized in this investigation were informative, indicating that they were appropriate for analyzing genetic diversity across and within the cowpea genotypes studied. The population structure was determined using two methodologies (STRUCTURE and DAPC), with the STRUCTURE analysis revealing the presence of six major populations (K = 6) and the DAPC analysis revealing five significant clusters (K = 5) for the 62 cowpea genotypes.
The high fixation index (Fst) value among genotypes between clusters verified the observed genetic differentiation. This contrasts with studies on cowpea by Fatokun et al. (2018) and cassava by Rabbi et al. (2015), which both showed a low fixation index. The current study’s low projected heterozygosity may be ascribed to the genotypes’ complicated breeding history in selected geographical origins. Even though the genetic variants discovered in this study were low, it provided vital information that might be used in breeding and research in Uganda. The germplasm may be examined for characteristics and used to drive genotype selection for genomic prediction and genome-wide association studies.
Conclusion
Our research found significant genetic heterogeneity in cowpea genotypes from Uganda, Nigeria, Ghana, Kenya, and the United States. SNP markers obtained from DArT-Seq were useful in evaluating genetic diversity and relatedness among parental cowpea lines used at the breeding program at MaRCCI, Uganda. The findings of this molecular characterization might be useful in Ugandan cowpea improvement efforts, serving as a guide for selecting accessions with desired features for breeding reasons. The genetic diversity discovered in the germplasm would be beneficial to the country’s cowpea development effort. The genetic diversity of the parental lines assembled for cowpea breeding in Uganda is a crucial aspect of the breeding program as it will provide the genetic material for developing improved cultivars with desired traits such as high yield, disease resistance, and adaptability to local growing conditions.
Data Availability
All the data and plant material are available from the corresponding author.
References
Acquaah G (2012) Principles of plant genetics and breeding. Second ed. (2nd ed.). John Willey and Sons Ltd, UK
Adu BG, Akromah R, Amoah S, Nyadanu D, Yeboah A, Aboagye LM, Amoah RA, Owusu EG (2021) High-density DArT-based SilicoDArT and SNP markers for genetic diversity and population structure studies in cassava (Manihot esculenta Crantz). Plos One16(7). https://doi.org/10.1371/journal.pone.0255290
Agre P, Asibe F, Darkwa K, Edemodu A, Bauchet G, Asiedu R, Adebola P, Asfaw A (2019) Phenotypic and molecular assessment of genetic structure and diversity in a panel of winged yam (Dioscorea alata) clones and cultivars. Sci Rep 9(1):18221. https://doi.org/10.1038/s41598-019-54761-3
Amponsah AE, Esuma W, Alicai T, Bhattacharjee R, Dramadri IO, Edema R, Chamba EB, Odong TL (2023) Genetic diversity and population structure of Uganda’s yam (Dioscorea spp.) genetic resource based on DArTseq. Plos One18(2):e0277537. https://doi.org/10.1371/journal.pone.0277537
Asiedu RN, Bai K, Ekanayake I, Wanyera N (1998) Genetic improvement. In Food Yams: Advances in research. (pp. 63–104)
Ayesiga SB, Rubaihayo P, Oloka BM, Dramadri IO, Edema R, Sserumaga JP (2022) Genetic variation among tropical maize inbred lines from NARS and CGIAR breeding programs. Plant Mol Biol Report. https://doi.org/10.1007/s11105-022-01358-2
Bado BV, Bationo A, Cescas MP (2006) Assessment of cowpea and groundnut contributions to soil fertility and succeeding sorghum yields in the Guinean savannah zone of Burkina Faso (West Africa). Biol Fertil Soils 43(2):171–176. https://doi.org/10.1007/s00374-006-0076-7
Bhattacharjee R, Agre P, Bauchet G, de Koeyer D, Lopez-Montes A, Kumar PL, Abberton M, Adebola P, Asfaw A, Asiedu R (2020) Genotyping-by-sequencing to unlock genetic diversity and population structure in white yam (Dioscorea rotundata Poir.). Agronomy 10(9):1437. https://doi.org/10.3390/agronomy10091437
Blyton MDJ, Flanagan NS (2006) A comprehensive guide to GenAlEx 6.5. (6.5). Australian National University
Boukar O, Belko N, Chamarthi S, Togola A, Batieno J, Owusu E, Haruna M, Diallo S, Umar ML, Olufajo O, Fatokun C (2019) Cowpea (Vigna unguiculata): genetics, genomics, and breeding. Plant Breeding 138(4):415–424. https://doi.org/10.1111/pbr.12589
Boukar O, Fatokun CA, Huynh B-L, Roberts PA, Close TJ (2016) Genomic tools in cowpea breeding programs: status and perspectives. Frontiers in Plant Science7. https://doi.org/10.3389/fpls.2016.00757
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics23(19). https://doi.org/10.1093/bioinformatics/btm308
Carvalho M, Lino-Neto T, Rosa E, Carnide V (2017) Cowpea: a legume crop for a challenging environment. J Sci Food Agric 97(13):4273–4284. https://doi.org/10.1002/jsfa.8250
Chen H, Chen H, Hu L, Wang L, Wang S, Wang ML, Cheng X (2017) Genetic diversity and a population structure analysis of accessions in the Chinese cowpea [ Vigna unguiculata (L.) Walp.] germplasm collection. The Crop Journal5(5), 363–372. https://doi.org/10.1016/j.cj.2017.04.002
Chidebe IN, Jaiswal SK, Dakora FD (2018) Distribution and phylogeny of microsymbionts associated with cowpea (Vigna unguiculata) nodulation in three agroecological regions of Mozambique. Appl Environ Microbiol84(2). https://doi.org/10.1128/AEM.01712-17
Damarany A, El-Dkeshy MHZ, Attallah SY, Haridy AG (2018) Molecular markers and genetic variation revealed by RAPD-PCR in seven cowpea (Vigna unguiculata (L.) Walp.) cultivars. Int J Biosci 13:66–74
Desalegne BA, Mohammed S, Dagne K, Timko MP (2016) Assessment of genetic diversity in Ethiopian cowpea [Vigna unguiculata (L.) Walp.] germplasm using simple sequence repeat markers. Plant Mole Biol Rep34(5):978–992. https://doi.org/10.1007/s11105-016-0979-x
Dias FTC, Bertini CHCM, Silva APM, Cavalcanti JJV (2015) Genetic variability in early-cycle erect cowpea analysed with RAPD and ISSR markers. Revista Ciência Agronomica 46:563–572
Earl DA, VonHoldt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Gen Resour4(2). https://doi.org/10.1007/s12686-011-9548-7
Eltaher S, Sallam A, Belamkar V, Emara HA, Nower AA, Salem KFM, Poland J, Baenziger P S (2018) Genetic diversity and population structure of F3:6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front Genet 9. https://doi.org/10.3389/fgene.2018.00076
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fatokun C, Girma G, Abberton M, Gedil M, Unachukwu N, Oyatomi O, Yusuf M, Rabbi I, Boukar O (2018) Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci Rep 8(1): 16035. https://doi.org/10.1038/s41598-018-34555-9
Gbedevi KM, Boukar O, Ishikawa H, Abe A, Ongom PO, Unachukwu N, Rabbi I, Fatokun C (2021) Genetic diversity and population structure of cowpea [Vigna unguiculata (L.) Walp.] germplasm collected from Togo based on DArT markers. Genes12(9):1451. https://doi.org/10.3390/genes12091451
Ghalmi N, Malice M, Jacquemin, J-M, Ounane S-M, Mekliche L, Baudoin J-P (2010) Morphological and molecular diversity within Algerian cowpea (Vigna unguiculata (L.) Walp.) landraces. Gen Resour Crop Evol 57(3):371–386. https://doi.org/10.1007/s10722-009-9476-5
Gomes A, Nhantumbo N, Ferreira-Pinto M, Massinga RC, Ramalho J, Ribeiro-Barros A (2019) Breeding elite cowpea [ Vigna unguiculata (L.) Walp] varieties for improved food security and income in Africa: opportunities and challenges. In Legume Crops - Characterization and Breeding for Improved Food Security. Intech Open. https://doi.org/10.5772/intechopen.84985
Gonçalves A, Goufo P, Barros A, Domínguez-Perles R, Trindade H, Rosa EAS, Ferreira L, Rodrigues M (2016) Cowpea (Vigna unguiculata L. Walp), a renewed multipurpose crop for a more sustainable agri-food system: nutritional advantages and constraints. J Sci Food Agri96(9):2941–2951. https://doi.org/10.1002/jsfa.7644
Grzebelus D, Iorizzo M, Senalik D, Ellison S, Cavagnaro P, Macko-Podgorni A, Heller-Uszynska K, Kilian A, Nothnagel T, Allender C, Simon PW, Baranski R (2014) Diversity, genetic mapping, and signatures of domestication in the carrot (Daucus carota L.) genome, as revealed by Diversity Arrays Technology (DArT) markers. Mole Breed 33(3):625–637. https://doi.org/10.1007/s11032-013-9979-9
Hall AE (2012) Phenotyping cowpeas for adaptation to drought. Front Physiol 3. https://doi.org/10.3389/fphys.2012.00155
Huynh B, Close TJ, Roberts PA, Hu Z, Wanamaker S, Lucas MR, Chiulele R, Cissé N, David A, Hearne S, Fatokun C, Diop NN, Ehlers JD (2013) Gene pools and the genetic architecture of domesticated cowpea. The Plant Genome 6(3). https://doi.org/10.3835/plantgenome2013.03.0005
Jayathilake C, Visvanathan R, Deen A, Bangamuwage R, Jayawardana BC, Nammi S, Liyanage R (2018) Cowpea: an overview of its nutritional facts and health benefits. J Sci Food Agric 98(13):4793–4806. https://doi.org/10.1002/jsfa.9074
Ketema S, Tesfaye B, Keneni G, Amsalu Fenta B, Assefa E, Greliche N, Machuka E, Yao N (2020) DArTSeq SNP-based markers revealed high genetic diversity and structured population in Ethiopian cowpea [Vigna unguiculata (L.) Walp] germplasms. Plos One 15(10):e0239122. https://doi.org/10.1371/journal.pone.0239122
Kilian A, Sanewski G, Ko L (2016) The application of DArTseq technology to pineapple. Acta Hortic 1111:181–188
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S (2012) SNP markers and their impact on plant breeding. International Journal of Plant Genomics 2012:1–11. https://doi.org/10.1155/2012/728398
Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo M, Alhakami H, Alpert M, Atokple I, Batieno BJ, Boukar O, Bozdag S, Cisse N, Drabo I, Ehlers JD, Farmer A, Fatokun C, Gu YQ, Guo Y, Huynh B, Close TJ (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. The Plant Journal 89(5):1042–1054. https://doi.org/10.1111/tpj.13404
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32(2):261–285. https://doi.org/10.1080/13102818.2017.1400401
Ndjiondjop M-N, Semagn K, Gouda AC, Kpeki SB, Dro Tia D, Sow M, Goungoulou A, Sie M, Perrier X, Ghesquiere A, Warburton ML (2017) Genetic variation and population structure of Oryza glaberrima and development of a mini-core collection using DArTseq. Front Plant Sci 8. https://doi.org/10.3389/fpls.2017.01748
Olufisayo AK, Mercy OO, Iyiola F (2016) Characterization of mutant cowpea [Vigna unguiculata (L) Walp] lines using random amplified polymorphic DNAs (RAPDs) and amplified fragment length polymorphism (AFLP) markers. Afr J Biotech 15(45):2530–2537. https://doi.org/10.5897/AJB2015.14539
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 28(19). https://doi.org/10.1093/bioinformatics/bts460
Porras-Hurtado L, Ruiz Y, Santos C, Phillips C, Carracedo Á, Lareu MV (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet 4. https://doi.org/10.3389/fgene.2013.00098
R Core Team (2020) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/
Rabbi IY, Kulakow PA, Manu-Aduening JA, Dankyi AA, Asibuo JY, Parkes EY, Abdoulaye T, Girma G, Gedil MA, Ramu P, Reyes B, Maredia MK (2015) Tracking crop varieties using genotyping-by-sequencing markers: a case study using cassava (Manihot esculenta Crantz). BMC Genet16(1). https://doi.org/10.1186/s12863-015-0273-1
Ragul S, Manivannan N, Mahalingam A, Rajendra Prasad VB, Lakshmi Narayanan S (2018) SSR marker based DNA fingerprinting for cowpea varieties of Tamil Nadu [Vigna unguiculata (L.) Walp.]. Int J Curr Microbiol Appl Sci 7(04):641–647. https://doi.org/10.20546/ijcmas.2018.704.072
Reyes-Valdés MH, Santacruz-Varela A, Martínez O, Simpson J, Hayano-Kanashiro C, Cortés-Romero C (2013) Analysis and optimization of bulk DNA sampling with binary scoring for germplasm characterization. PLoS One 8(11). https://doi.org/10.1371/journal.pone.0079936
Seo E, Kim K, Jun T-H, Choi J, Kim S-H, Muñoz-Amatriaín M, Sun H, Ha B-K (2020) Population structure and genetic diversity in Korean cowpea germplasm based on SNP markers. Plants 9(9):1190. https://doi.org/10.3390/plants9091190
Talabi AO, Badu-Apraku B, Fakorede MAB (2017) Genetic variances and relationship among traits of an early maturing maize population under drought-stress and low nitrogen environments. Crop Sci 57(2):681–692. https://doi.org/10.2135/cropsci2016.03.0177
Teeken B, Olaosebikan O, Haleegoah J, Oladejo E, Madu T, Bello A, Parkes E, Egesi C, Kulakow P, Kirscht H, Tufan HA (2018) Cassava trait preferences of men and women farmers in Nigeria: implications for breeding. Econ Bot 72(2):263–277
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated, and elite barleys. Plant Sci 173(6):638–649. https://doi.org/10.1016/j.plantsci.2007.08.010
Wamalwa EN, Muoma J, Wekesa C (2016) Genetic diversity of cowpea (Vigna unguiculata (L.) Walp.) accession in Kenya gene bank based on simple sequence repeat markers. Int J Genom 2016:1–5. https://doi.org/10.1155/2016/8956412
Wang X, Chang L, Tong Z, Wang D, Yin Q, Wang D, Jin X, Yang Q, Wang L, Sun Y (2016) Proteomics profiling reveals carbohydrate metabolic enzymes and 14–3–3 proteins play important roles for starch accumulation during cassava root tuberization. Sci Rep 6(19643)
Xia L, Peng K, Yang S, Wenzl P, Carmen de Vicente M, Fregene M, Kilian A (2005) DArT for high-throughput genotyping of cassava (Manihot esculenta) and its wild relatives. Theo Appl Genet 110(6). https://doi.org/10.1007/s00122-005-1937-4
Xiong H, Shi A, Mou B, Qin J, Motes D, Lu W, Ma J, Weng Y, Yang W, Wu D (2016) Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). Plos One11(8):e0160941. https://doi.org/10.1371/journal.pone.0160941
Acknowledgements
We acknowledge support and resources from the staff and management of Makerere Regional Center for Crop Improvement (MaRCCI) and the Department of Agricultural Science of Makerere University.
Funding
This study was supported by funds from the World Bank project through Makerere Regional Center for Crop Improvement (MaRCCI) (Grant: IDA Credit No. 57970 UG).
Author information
Authors and Affiliations
Contributions
Conceptualization, R.E, I.O.D, and E.A.A; methodology, I.O.D and E.A.A; formal analysis, E.A.A and I.O.D; investigation, R.E, I.O.D, and E.A.A; data curation, I.O.D and E.A.A; writing—original draft preparation, E.A.A.; writing—review and editing, R.E, A.A.O, S.M.T, A.B, and A.I; supervision, I.O.D and R.E; funding acquisition, R.E and I.O.D. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Edema, R., Adjei, E.A., Ozimati, A.A. et al. Genetic Diversity of Cowpea Parental Lines Assembled for Breeding in Uganda. Plant Mol Biol Rep 41, 713–725 (2023). https://doi.org/10.1007/s11105-023-01394-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-023-01394-6