Abstract
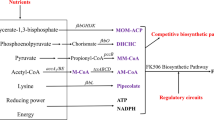
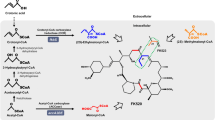
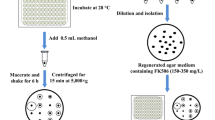
FK506 is a clinically important macrocyclic polyketide with immunosuppressive activity produced by Streptomyces tsukubaensis. However, the low titer at which it is produced is a bottleneck to its application and use in industrial processes. We have overexpressed five potential targets associated with FK506 production (fkbO, fkbL, fkbP, fkbM, fkbD) which were identified in our previous study, with the aim to improve FK506 production. The results of the analysis showed that the constructed strains with an additional copy of each gene increased FK506 production by approximately 10–40 % compared with the wild-type strain D852. The results of the gene expression analysis indicated that each gene was upregulated. Combinatorial overexpression of the five genes resulted in a 146 % increase in the FK506 titer to 353.2 mg/L, in comparison with the titer produced by D852. To further improve the production of FK506 by the engineered strain HT-FKBOPLMD, we supplemented the medium with various nutrients, including soybean oil, lactate, succinate, shikimate, chorismate, lysine, pipecolate, isoleucine and valine. Optimization of feeding concentrations and times resulted in HT-FKBOPLMD being able to produce approximately 70 % more FK506, thereby reaching the maximal titer of 457.5 mg/L, with lower amounts of by-products (FK520 and 37,38-dihydro-FK506). These results demonstrate that the combination of the metabolically engineered secondary pathways and the exogenous feeding strategies developed here was able to be successfully applied to improve the production of industrially and clinically important compounds.








Similar content being viewed by others
References
Alper H, Jin YS, Moxley JF, Stephanopoulos G (2005) Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7(3):155–164. doi:10.1016/j.ymben.2004.12.003
Andexer JN, Kendrew SG, Nur-e-Alam M, Lazos O, Foster TA, Zimmermann AS, Warneck TD, Suthar D, Coates NJ, Koehn FE, Skotnicki JS, Carter GT, Gregory MA, Martin CJ, Moss SJ, Leadlay PF, Wilkinson B (2011) Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc Natl Acad Sci USA 108(12):4776–4781. doi:10.1073/pnas.1015773108
Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J (2009) Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng 11(6):328–334. doi:10.1016/j.ymben.2009.07.001
Chen D, Zhang Q, Zhang Q, Cen P, Xu Z, Liu W (2012) Improvement of FK506 production in Streptomyces tsukubaensis by genetic enhancement of the supply of unusual polyketide extender units via utilization of two distinct site-specific recombination systems. Appl Environ Microbiol 78(15):5093–5103. doi:10.1128/aem.00450-12
Cheng YR, Fang A, Demain AL (1995) Effect of amino acids on rapamycin biosynthesis by Streptomyces hygroscopicus. Appl Microbiol Biotechnol 43(6):1096–1098
Choi HS, Lee SY, Kim TY, Woo HM (2010) In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol 76(10):3097–3105. doi:10.1128/aem.00115-10
Duan Y, Chen T, Chen X, Zhao X (2010) Overexpression of glucose-6-phosphate dehydrogenase enhances riboflavin production in Bacillus subtilis. Appl Microbiol Biotechnol 85(6):1907–1914. doi:10.1007/s00253-009-2247-6
DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28(3):350–356. doi:10.1021/ac60111a017
Fang A, Demain AL (1995) Exogenous shikimic acid stimulates rapamycin biosynthesis in Streptomyces hygroscopicus. Folia Microbiol 40(6):607–610
Görisch H, Lingens F (1973) Chorismate mutase from Streptomyces aureofaciens: a heat-stable enzyme. J Bacteriol 114(2):645–651
Gao H, Zhuo Y, Ashforth E, Zhang L (2010) Engineering of a genome-reduced host: practical application of synthetic biology in the overproduction of desired secondary metabolites. Protein Cell 1(7):621–626. doi:10.1007/s13238-010-0073-3
Gatto GJ, Boyne MT, Kelleher NL, Walsh CT (2006) Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J Am Chem Soc 128(11):3838–3847. doi:10.1021/ja0587603
Goranovic D, Kosec G, Mrak P, Fujs S, Horvat J, Kuscer E, Kopitar G, Petkovic H (2010) Origin of the allyl group in FK506 biosynthesis. J Biol Chem 285:14292–14300. doi:10.1074/jbc.M109.059600
Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R (2002) Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies. J Bacteriol 184(7):2019–2029. doi:10.1128/jb.184.7.2019-2029.2002
Huang D, Jia X, Wen J, Wang G, Yu G, Caiyin Q, Chen Y (2011) Metabolic flux analysis and principal nodes identification for daptomycin production improvement by Streptomyces roseosporus. Appl Biochem Biotechnol 165(7):1725–1739. doi:10.1007/s12010-011-9390-0
Huang D, Wen J, Wang G, Yu G, Jia X, Chen Y (2012) In silico aided metabolic engineering of Streptomyces roseosporus for daptomycin yield improvement. Appl Microbiol Biotechnol 94(3):637–649. doi:10.1007/s00253-011-3773-6
Jin YS, Stephanopoulos G (2007) Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab Eng 9(4):337–347. doi:10.1016/j.ymben.2007.03.003
König A, Schwecke T, Molnár I, Böhm GA, Lowden PAS, Staunton J, Leadlay PF (1997) The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin—nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces Hygroscopicus. Eur J Biochem 247(2):526–534. doi:10.1111/j.1432-1033.1997.00526.x
Khaw LE, Böhm GA, Metcalfe S, Staunton J, Leadlay PF (1998) Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J Bacteriol 180(4):809–814
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kino T, Hatanaka H, Hashimoto M, Nishiyama M, Goto T, Okuhara M, Kohsaka M, Aoki H, Imanaka H (1987) FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, and physico-chemical and biological characteristics. J Antibiot 40(9):1249–1255
Koju D, Maharjan S, Dhakal D, Yoo JC, Sohng JK (2012) Effect of different biosynthetic precursors on the production of nargenicin A1 from metabolically engineered Nocardia sp. CS682. J Microbiol Biotechnol 22(8):1127–1132. doi:10.4014/jmb.1202.02027
Kosec G, Goranovič D, Mrak P, Fujs Š, Kuščer E, Horvat J, Kopitar G, Petković H (2012) Novel chemobiosynthetic approach for exclusive production of FK506. Metab Eng 14(1):39–46. doi:10.1016/j.ymben.2011.11.003
Kudo F, Yonezawa T, Komatsubara A, Mizoue K, Eguchi T (2011) Cloning of the biosynthetic gene cluster for naphthoxanthene antibiotic FD-594 from Streptomyces sp. TA-0256. J Antibiot 64(1):123–132. doi:10.1038/ja.2010.145
Kuščer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petković H (2007) Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol 189(13):4756–4763. doi:10.1128/jb.00129-07
Maharjan S, Park JW, Yoon YJ, Lee HC, Sohng JK (2010) Metabolic engineering of Streptomyces venezuelae for malonyl-CoA biosynthesis to enhance heterologous production of polyketides. Biotechnol Lett 32(2):277–282. doi:10.1007/s10529-009-0152-9
Medema MH, Alam MT, Breitling R, Takano E (2011) The future of industrial antibiotic production: from random mutagenesis to synthetic biology. Bioeng Bugs 2(4):230–233. doi:10.1111/j.1751-7915.2010.00226.x
Mendes MV, Tunca S, Antón N, Recio E, Sola-Landa A, Aparicio JF, Martín JF (2007) The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab Eng 9(2):217–227. doi:10.1016/j.ymben.2006.10.003
Mo S, Ban YH, Park JW, Yoo YJ, Yoon YJ (2009) Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J Ind Microbiol Biotechnol 36(12):1473–1482. doi:10.1007/s10295-009-0635-7
Mo S, Kim DH, Lee JH, Park JW, Basnet DB, Ban YH, Yoo YJ, Chen SW, Park SR, Choi EA, Kim E, Jin YY, Lee SK, Park JY, Liu Y, Lee MO, Lee KS, Kim SJ, Kim D, Park BC, Lee SG, Kwon HJ, Suh JW, Moore BS, Lim SK, Yoon YJ (2011) Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J Am Chem Soc 133(4):976–985. doi:10.1021/ja108399b
Motamedi H, Shafiee A (1998) The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur J Biochem 256(3):528–534. doi:10.1046/j.1432-1327.1998.2560528.x
Motamedi H, Shafiee A, Cai SJ, Streicher SL, Arison BH, Miller RR (1996) Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J Bacteriol 178(17):5243–5248
Mouslim J, David L, Pétel G, Gendraud M (1993) Effect of exogeneous methyl oleate on the time course of some parameters of Streptomyces hygroscopicus NRRL B-1865 culture. Appl Microbiol Biotechnol 39(4–5):585–588. doi:10.1007/bf00205056
Murrell JM, Liu W, Shen B (2004) Biochemical characterization of the SgcA1 α-d-glucopyranosyl-1-phosphate thymidylyltransferase from the enediyne antitumor antibiotic C-1027 biosynthetic pathway and overexpression of sgcA1 in Streptomyces globisporus to improve C-1027 production. J Nat Prod 67(2):206–213. doi:10.1021/np0340403
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10(5):281–292. doi:10.1016/j.ymben.2008.07.001
Paiva NL, Demain AL, Roberts MF (1993) The immediate precursor of the nitrogen-containing ring of rapamycin is free pipecolic acid. Enzyme Microb Technol 15(7):581–585. doi:10.1016/0141-0229(93)90020-3
Parsons WH, Sigal NH, Wyvratt MJ (1993) FK-506–a novel immunosuppressant. Ann N Y Acad Sci 685(1):22–36. doi:10.1111/j.1749-6632.1993.tb35847.x
Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM (2006) Effects of methylmalonyl-CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. J Ind Microbiol Biotechnol 33(7):600–609. doi:10.1007/s10295-006-0094-3
Reeves AR, Cernota WH, Brikun IA, Wesley RK, Weber JM (2004) Engineering precursor flow for increased erythromycin production in Aeromicrobium erythreum. Metab Eng 6(4):300–312. doi:10.1016/j.ymben.2004.03.003
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16(3):313–340. doi:10.1099/00207713-16-3-313
Sierra-Paredes G, Sierra-Marcuño G (2008) Ascomycin and FK506: pharmacology and therapeutic potential as anticonvulsants and neuroprotectants. CNS Neurosci Ther 14(1):36–46. doi:10.1111/j.1527-3458.2008.00036.x
Singh BP, Behera BK (2009) Regulation of tacrolimus production by altering primary source of carbons and amino acids. Lett Appl Microbiol 49(2):254–259. doi:10.1111/j.1472-765X.2009.02652.x
Thykaer J, Nielsen J, Wohlleben W, Weber T, Gutknecht M, Lantz AE, Stegmann E (2010) Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab Eng 12(5):455–461. doi:10.1016/j.ymben.2010.05.001
Turło J, Gajzlerska W, Klimaszewska M, Król M, Dawidowski M, Gutkowska B (2012) Enhancement of tacrolimus productivity in Streptomyces tsukubaensis by the use of novel precursors for biosynthesis. Enzyme Microb Technol 51(6–7):388–395. doi:10.1016/j.enzmictec.2012.08.008
Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Bohm I, Mironenko T, Deacon M, Wheatcroft M, Wirtz G, Staunton J, Leadlay PF (2002) Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol Biotechnol 4(4):417–426
Yepes A, Rico S, Rodriguez-Garcia A, Santamaria RI, Diaz M (2011) Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PLoS ONE 6(5):e19980. doi:10.1371/journal.pone.0019980
Yin H, Xiang S, Zheng J, Fan K, Yu T, Yang X, Peng Y, Wang H, Feng D, Luo Y, Bai H, Yang K (2012) Induction of holomycin production and complex metabolic changes by the argR mutation in Streptomyces clavuligerus NP1. Appl Environ Microbiol 78(9):3431–3441. doi:10.1128/aem.07699-11
Yoon YJ, Choi CY (1997) Nutrient effects on FK-506, a new immunosuppressant, production by Streptomyces sp. in a defined medium. J Ferment Bioeng 83(6):599–603. doi:10.1016/s0922-338x(97)81145-2
Zhu X, Zhang W, Chen X, Wu H, Duan Y, Xu Z (2010) Generation of high rapamycin producing strain via rational metabolic pathway-based mutagenesis and further titer improvement with fed-batch bioprocess optimization. Biotechnol Bioeng 107(3):506–515. doi:10.1002/bit.22819
Acknowledgments
This work was financially supported by the National 973 Project of China (No. 2013CB733600), the Key Program of National Natural Science Foundation of China (No. 21236005) and the Program of Introducing Talents of Discipline to Universities (No. B06006). The authors especially thank Professor A.L. Demain in Drew University for valuable suggestion on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, D., Xia, M., Li, S. et al. Enhancement of FK506 production by engineering secondary pathways of Streptomyces tsukubaensis and exogenous feeding strategies. J Ind Microbiol Biotechnol 40, 1023–1037 (2013). https://doi.org/10.1007/s10295-013-1301-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1301-7