Abstract
Background
Flag leaf is the most essential organ for photosynthesis in rice and its size plays an important role in rice breeding for ideal plant-type. Flag leaf size affect photosynthesis to a certain extent, thereby influencing rice production. Several genes controlling leaf size and shape have been identified with mutants. Although a number of quantitative trait loci (QTLs) for leaf size and shape have been detected on 12 chromosomes with different populations of rice, few of them were cloned.
Results
The pair-wise correlation analysis was conducted on length, width and length-width ratio of the flag leaf, and yield per plant in the core recombinant inbred lines of Liang-You-Pei-Jiu (LYP9) developed in Hainan and Hangzhou. There were significant correlations among the three flag leaf size and shape traits. Interestingly, a positive correlation was found between flag leaf width and yield per plant. Based on the high-resolution linkage map we constructed before, 43 QTLs were detected for three flag leaf size and shape traits and yield per plant, among which 31 QTLs were unreported so far. Seven QTLs were identified common in two environments. And qFLW7.2, a new major QTL for flag leaf width, was fine mapped within 27.1 kb region on chromosome 7. Both qFLW7.2 and qPY7 were located in the interval of 45.30 ~ 53.34 cM on chromosome 7, which coincided with the relationship between yield per plant (PY) and flag leaf width (FLW).
Conclusion
qFLW7.2, which explained 14% of the phenotypic variation, increased flag leaf width with 93–11 allele. Two candidate genes were selected based on sequence variation and expression difference between two parents, which facilitated further QTL cloning and molecular breeding in super rice.
Similar content being viewed by others
Background
Rice is not only one of the most important food crops in China, but a staple food for more than half the world's population (Delseny et al. 2001). With increasing population, high yield has become one of targets in rice breeding. Photosynthesis is the primary source of grain yield in rice (Chen et al. 1995). The top three leaves of rice, particularly the flag leaf, are the main source of carbohydrates production (Abrol et al. 1993; Foyer, 1987). At least 50% of photosynthetic products for grain are provided by flag leaf, the most important organ for photosynthesis (Li et al. 1998). Some traits, such as size and shape of flag leaf, affect photosynthesis to a certain extent, thereby influencing production (Yue et al. 2006). Therefore, flag leaf shape is an index for ideal plant-type in rice breeding (Yang and Yang 1998; Yuan, 1997; Zhou et al. 1995).
Besides several genes controlling leaf size and shape cloned with mutants (Fujino et al. 2008; Qi et al. 2008; Zhang et al. 2009; Hu et al. 2010; Xiang et al. 2012), some QTLs for the traits of flag leaf size and rice yield have also been mapped with diverse populations, such as F2, doubled haploid (DH) and recombinant inbred lines (RILs) (Wang et al. 2004; Peng et al. 2007; Wang et al. 2009; Jiang et al. 2010a). Yan and Wang (1990) studied 11 flag leaf traits in indica-japonica hybrids, and argued that flag leaf length (FLL), FLW and flag leaf area (FLA) were controlled by two pairs of genes with at least more than 60% heritability. In recent years, with the rapid development of molecular markers and the increase in resolution of the linkage map, numbers of QTLs for flag leaf size and shape have been reported in rice. Li et al. (2000) detected 13 QTLs for FLL, FLW, FLA and length-width ratio (LWR), explained 8.7% ~ 18.5% of phenotypic variation, with DH population from a cross of Zhaiye Qing 8 and Jingxi 17. Using a DH population and a genetic map with 175 SSR markers under multi environments, Cao et al. (2007) detected 15 QTLs affected FLL, whose genetic intervals were 2 ~ 18 cM. Xiao et al. (2007) also identified 8 QTLs for the traits of FLL, FLW and FLA in the backcross recombinant inbred lines (BILs) derived from a cross between Koshihikari and Kasalath. However, most studies focused on the size and shape of the flag leaf and few involved in their relationship with yield. And so far, no QTL for flag leaf size has been isolated yet.
In the study, the relationship between flag leaf size and PY were analyzed. QTLs for three flag leaf traits and yield per plant (PY) were mapped based on a high-density linkage map by resequencing the parents of LYP9 and 132 core RILs (Gao et al., 2013). A novel major QTL for flag leaf width was fine mapped and 2 candidate genes were selected, by which providing a basis for further cloning of the QTL and improvement of ideal plant-type in hybrid rice.
Results
Phenotypic variation of the parents and RILs
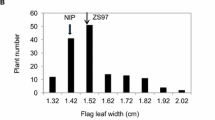
The phenotypic differences between 93–11 and PA64s are displayed and summarized in Figure 1A and Table 1. The t-test revealed that the differences between two parents were extremely significant concerning FLW and PY in Hangzhou, as well as for FLL, FLW and PY in Hainan. While for FLL and LWR in Hangzhou, LWR in Hainan, the differences were significant between the parents. Microscopic observation with flag leaves indicated difference in the number of small veins on average between the parents, although little difference in the number of large veins (Figure 1B, C, D, E and Table 2). Continuous distributions were observed in the RIL population for all four traits and the average value of each trait was close to its mid-parent value (Figure 2), indicating all of the four traits were quantitative traits controlled by multi-genes and satisfied the demands for QTL analysis.
Correlation analysis of the four traits
The correlations among the four characteristics were shown in Table 3. The results showed that significant positive correlations were detected between PY and FLW in both Hainan and Hangzhou. Meanwhile, FLL was positively correlated in extreme significance with FLW and LWR. Reasonably, negative correlations were identified between FLW and LWR in both Hainan and Hangzhou.
QTL detection for flag leaf size and yield per plant
A total of 43 QTLs were detected for the traits of FLL, FLW, LWR and PY in both Hainan and Hangzhou, distributing on 10 chromosomes except for chromosome 2 and chromosome 9 (Table 4; Figure 3). Nine QTLs for FLL were identified, including 4 QTLs in Hainan and 5 QTLs in Hangzhou, each explained 4% ~ 11% of phenotypic variation. Fourteen QTLs for FLW were detected and each QTL explained 4% ~ 24% of phenotypic variation. In Hangzhou, the 93–11 alleles increased FLW at qFLW8 and qFLW7.2, explained 24% and 17% of the phenotypic variation, respectively. In Hainan, the 93–11 alleles also increased FLW at qFLW1 and qFLW7.2 which explained 15% and 14% of the phenotypic variation, respectively. For the trait of LWR, 12 QTLs were detected including 5 QTLs in Hainan and 7 QTLs in Hangzhou. Eight QTLs were identified for PY in both Hainan and Hangzhou. In Hangzhou, the 93–11 allele increased PY at qPY7 which can explain 10% of the phenotypic variation and located within 49.20 ~ 53.34 cM on chromosome 7.
Among all the 43 QTLs detected with RILs, 7 QTLs were commonly identified in both Hainan and Hangzhou, demonstrating their environmental independent. Five of them distributing on chromosome 4, 5, 7, 7 and 12 were responsible for FLW and the other two QTLs on chromosome 7 and 11 for LWR. There were 10 clusters involving at least two QTLs, among which three clusters on chromosome 7, 8 and 12 simultaneously responsible for FLW and PY, which coincided with significant correlations between the two traits.
Fine mapping and candidate gene analysis of a major QTL qFLW7.2
Among 43 QTLs detected in RILs, 31 QTLs were unreported so far, including qFLW7.2 identified in both Hainan and Hangzhou. For fine mapping of the new major QTL, residual heterozygous line (RHL) were selected from a large RIL population, carrying approximately 484 kb heterozygous segment on the long arm of chromosome 7. Then phenotypic character was measured in F2 population including 1520 individuals derived from the RHL. Three insertion-deletion (InDel) and five single nucleotide polymorphism (SNP) markers were developed by comparing the sequences of the parents. Combining the genotype and phenotype of individuals, the QTL was delimited between two InDel markers INDEL7-2 and INDEL7-3 in 27.1 kb interval (Figure 4B). The target region contains 3 predicted genes (LOC_Os07g41180, LOC_Os07g41190 and LOC_Os07g41200) based on Rice Genome Annotation Project Website (http://rice.plantbiology.msu.edu/). Sequence variations of those genes between two parents were identified and expressions at RNA level were analyzed in leaves of the parents at booting stage (Figure 4C; Figure 5). Four SNPs causing amino acid change and 3 SNPs existed in exons and the promoter region, respectively in LOC_Os07g41180 gene. And the gene LOC_Os07g41200 had 2 nonsynonymous SNPs in one exon, 3 SNPs and an InDel in the promoter (Figure 4C). Both genes expressed at significantly different level in PA64s and two NILs (NIL-PA64s-1 and NIL-PA64s-2) compared with 93–11 (Figure 5). There were only 6 SNPs in the promoter of LOC_Os07g41190 gene and no significantly different expression in PA64s and two NILs compared with 93–11. Therefore, LOC_Os07g41180 and LOC_Os07g41200 were selected candidates for qFLW7.2.
Fine mapping of qFLW7.2 for FLW. A. Distribution of FLW in the F2 population derived from RHL. B. qFLW7.2 was narrowed down to a 27.4 kb interval defined by markers INDEL7-2 and INDEL 7–3. Values represent means ± SD. Gray represents heterotype. The superscript letters (a, b and c) indicate significant differences in the trait of the recombinants compared with two parents at a level of 0.01. C. Structure and mutated sites of two candidate genes. Grey boxes represent exons.
Comparison of flag leaf size between the two near isogenic lines (NILs) and 93–11 revealed FLW was wider in 93–11 than in NIL-PA64s-1 and NIL-PA64s-2, while little difference found between 93–11 and two NILs in FLL (Figure 6A, B, C). Meanwhile, significant difference was observed between 93–11 and two NILs in PY (Figure 6D). It indicated that the allele from PA64s affect flag leaf width and yield per plant at qFLW7.2 between INDEL7-2 and INDEL7-3, approximately 27.1 kb physical distance (Figure 6A).
Comparison in FLL, FLW and PY of NIL-PA64s-1, NIL-PA64s-2 and 93-11. A. Schematic graph of chromosomes of NIL-PA64s-1 and NIL-PA64s-2. B, C, D. Comparison of FLL, FLW and PY between NIL-PA64s-1, NIL-PA64s-2 and 93–11. NIL-PA64s-1 and NIL-PA64s-2, carrying homozygous alleles of PA64s in the target QTL region (black box) between INDEL7-1 and INDEL7-3, INDEL7-2 and RM234, respectively, were developed from one CSSL with 93–11 background. Values represent means ± SD of three plants.
Discussion
Leaf is the main organ for photosynthesis in rice. Several rice mutants for leaf size and shape have been identified and some corresponding genes have been cloned. Fujino et al. (2008) isolated a spontaneous mutant with narrow leaf, termed narrow leaf 7 (nal7). The gene Nal7, encoding a flavin-containing mono-oxygenase, were fine mapped on chromosome 3 and cloned with F2 population. The Nal1 gene located on chromosome 4, whose mutation affected lateral leaf growth and exhibited narrow leaf, encodes a plant specific protein of unknown biochemical function (Qi et al. 2008). And the NRL1 gene was fine mapped on the chromosome 12 and coded for the cellulose synthase-like protein D4 (Hu et al. 2010). Currently, many QTLs have been fine mapped related to flag leaf size and shape. Jiang et al. (2010b) detected 3 QTLs for FLL on chromosome 3, 6, and 9 using a separated population involving 176 individuals from a cross of Shennong 265/LTH. And qFLL9 was further mapped within a 198 kb interval on chromosome 9 by analyzing F2 population including 889 individuals derived from the RHL. Shen et al. (2012) fine mapped qFLL6.2 within 62.1 kb on the short arm of chromosome 6 by a F2 population derived from the RHL. Wang et al. (2011) narrowed the location of qFL1 for flag leaf length to a 31 kb region containing 4 predicted genes with BC2F3 and BC3F2. In our study, 43 QTLs were detected in two environments with the shortest genetic interval 0.19 cM in a high-density linkage map using the RIL population. Among 31 unreported QTL, a novel major QTL qFLW7.2, detected in both Hainan and Hangzhou was fine mapped in a 27.1 kb physical interval on chromosome 7 with the F2 population derived from a RHL. Two NILs containing the region from PA64s also showed narrower FLW compared with 93–11. Two candidate genes, LOC_Os07g41180 and LOC_Os07g41200, encoding RNA-binding protein and unknown expressed protein respectively, were selected based on sequence variations and transcriptional expression and to be further testified by complementation test.
As the most important and efficient functional leaf at grain filling stage, flag leaf shape is one of the essential traits for ideal plant-type in super rice breeding (Chen et al. 2001). It played an important role in molecular genetics and marker assisted selection (MAS) of flag leaf size and shape related traits. Here, PY and FLW were found significantly and positively correlated, which suggested that appropriate increase in FLW may raise PY correspondingly. Both qFLW7.2 and qPY7 were located in the interval of 45.30 ~ 53.34 cM on chromosome 7, which coincided with the relationship between PY and FLW. It suggested that qFLW7.2 may show pleictropism and play an important role in the formation of rice yield. Previous studies found that FLW was significantly correlated to panicle number and spikelet number per panicle (Zhou et al. 2012). Recent studies showed that SPIKE, a nal1 allele, can increase 13 ~ 36% of the yield of the NIL derived from indica variety IR64, with leaf area significantly increased. Therefore, SPIKE was believed to induce the enhancement of source size and translocation capacity as well as sink size (Fujita et al. 2013). Therefore, it was reasonable that wider flag leaf may increase photosynthetic area, so that the source supply was enhanced, and thereby rice yield improved. Moreover, molecular markers adjacent to qFLW7.2 can also be utilized effectively in controlling flag leaf width and high-yield breeding in rice.
Conclusion
In this study, using high-density SNP linkage map, 43 QTLs were detected in Hangzhou and Hainan to control rice leaf morphology and yield per plant. Owing to the increased precision and sensitivity of detection, minimum QTL interval reached 0.19 cM and 31 QTLs were novel. With the F2 population derived from a RHL, qFLW7.2, a new major QTL for FLW, was fine mapped within 27.1 kb physical interval on chromosome 7. Two candidate genes were finally selected based on difference in genomic sequence and transcriptional expression. Because the significantly positive correlation between FLW and PY, together with common interval shared by QTLs for FLW and PY, appropriate increase in FLW may raise PY correspondingly during molecular breeding for ideal plant-type in rice.
Methods
Mapping population and genetic map
The core mapping population of 132 LYP9 RILs was derived by single-seed descend from a cross between an elite paternal inbred Oryza sativa. indica cv. 93–11 and the maternal inbred Oryza sativa. javonica cv. Peiai 64 s (PA64s), a photo-thermo-sensitive male sterile line. The population was developed in the experimental fields at China National Rice Research Institute in Hangzhou, Zhejiang Province and in Lingshui, Hainan Province, China. After 12 generations of self-fertilization, genomic DNA samples of the F13 RILs were isolated for genotyping. High-density map of genome-wide graphic genotypes was constructed using single nucleotide polymorphism SNP markers as described previously (Gao et al., 2013). The RHL carrying approximately 484 kb heterozygous segment on the long arm of chromosome 7 was segregated from large high-generation RILs with 1520 individuals. A F2 population derived from the RHL was used for fine mapping. Two NILs carrying homozygous alleles of PA64s in the target QTL region between InDel markers INDEL7-1 and INDEL7-3, INDEL7-2 and a simple sequence repeat (SSR) marker RM234 (Tian et al., 2013), designated respectively NIL-PA64s-1 and NIL-PA64s-2, were also developed from one chromosome segment substitution line (CSSL) with 93–11 background (Table 5).
Field experiment and trait measurement
The 132 RILs and two parental lines were grown in Hangzhou in 2011 and Hainan in 2012. The F2 population derived from the RHL was grown in Hangzhou in 2013. NILs were cultivated in Hangzhou in 2014 following a randomized block design with three repeats. 25-day-old seedlings of each line were transplanted into a four-row plot with six plants per row and spacing of 15 cm × 25 cm. The field management followed normal agricultural practice.
Three flag leaf size and shape traits were investigated for four plants per line in the middle of rows 10 days after heading. The flag leaf length (FLL, cm) and flag leaf width (FLW, cm) were measured on three tillers. One derived trait, the length-width ratio (LWR) = FLL/FLW, was calculated. The trait yield per plant (PY, g) was also examined for the plants whose flag leaf size and shape had been investigated.
Leaf sections and microscopic analysis
Flag leaves of two parents were collected at heading stage and fixed in Formalin-Aceto-Alcohol (FAA). The samples were dehydrated through a graded ethanol series, then embedded in Paraffin (Surgipath®) and polymerized at 60°C. Finally, the materials were sectioned and stained with 1% toluidine blue before examination under an ECLIPSE 50i microscope (Nikon) (Hu et al. 2010).
QTL analysis
Phenotypic variations and correlations were analyzed by SAS 8.0 software. QTL analysis was performed with the R/qtl_1.26-14 (http://www.rqtl.org/) using Composite Interval Mapping (CIM). LOD threshold for each dataset was set based on a permutation test (1,000 permutation, P = 0.05). It was considered as a major effect QTL when its LOD score was larger than 2.5. PEV was estimated by ANOVA. QTLs were named according to McCouch et al. (1997).
Development of InDel and SNP markers for fine mapping
Primers were designed around qFLW7.2 on chromosome 7 on the basis of insertions/deletions (InDels) and SNPs identified between 93–11 and PA64s (Table 5). Genotypes of SNP markers were screened by high-resolution dissociation curve analysis system (LightScanner 96, Idaho Technology Inc.).
RNA extraction and real time PCR analysis
Total RNA was isolated from flag leaf at booting stage with RNA extraction kit (Axygen). DNase treatment, cDNA synthesis, primer design and SYBR Green I real time PCR were carried out as described (Vandesompele et al. 2002) using a Rever Tra Ace® qPCR-RT kit (TOYOBA, Japan). Real time PCR amplification mixtures (10 μl) contained 50 ng template cDNA, 2 × SYBR Green PCR Master Mix (Applied Biosystems), and 200 nM forward and reverse primers. Reactions were run on an ABI PRISM_7900HT Sequence Detector (Applied Biosystems). The relative expression level of each transcript was obtained by comparing to the expression of the OsActin1 gene. Primers for candidate genes and OsActin1 are listed in Table 6.
References
Abrol YP, Mohanty P, Govindjee A. Photosynthesis: photoreactions to plant productivity. Dordrecht: Kluwer Academic Publishers; 1993. p. 104–53.
Cao LY, Yang MG, Jun Z. QTL analysis for flag leaf length in rice DH population under multi environments. Yi Chuan Xue Bao. 2007;33:223–9.
Chen WF, Xu ZJ, Zhang LB. Physiological bases of super high yield breeding in rice. Shenyang: Liaoning Science and Technology Publishing Company; 1995. p. 1–2.
Chen WF, Xu ZJ, Zhang WZ, Zhang LB, Yang SR. Creation of New Plant Type and Breeding Rice for Super High Yield. Yi Chuan Xue Bao. 2001;27:665–72.
Delseny M, Salses J, Cooke R, Sallaud C, Regad F, Lagoda P, et al. Rice genomics: present and future. Plant Physiol Biochem. 2001;39:323–34.
Foyer C. The basis for source-sink interaction in leaves. Plant Physiol Biochem. 1987;25:649–57.
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics. 2008;279:499–507.
Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, et al. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci U S A. 2013;110:20431–6.
Gao ZY, Zhao SC, He WM, Guo LB, Peng YL, Wang JJ, et al. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci U S A. 2013;110:14492–7.
Hu J, Zhu L, Zeng DL, Gao ZY, Guo LB, Fang Y, et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol. 2010;73:283–92.
Jiang GH, Xu CG, Li XH, He YQ. Characterization of the genetic basis for yield and its component traits of rice revealed by doubled haploid population. Yi Chuan Xue Bao. 2004;31:63–72.
Jiang SK, Zhang XJ, Huang C, Xing YN, Xu ZJ, Chen WF. Comparison of genetic linkage map and QTLs controlling flag leaf traits based on F2 and F2:6 populations derived from japonica rice. Chin J Rice Sci. 2010a;24:372–8.
Jiang SK, Zhang XJ, Wang JY, Chen WF, Xu ZJ. Fine mapping of the quantitative trait locus qFLL9 controlling flag leaf length in rice. Euphytica. 2010b;176:341–7.
Li ZK, Pinson SR, Stansel JW, Paterson AH. Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (Oryza sativa L.). Mol Breeding. 1998;4:419–26.
Li SG, He P, Wang YP, Li HY, Chen Y, Zhou KD, et al. Genetic analysis and gene mapping of the leaf traits in rice (Oryza sativa L.). Acta Genetica Sin. 2000;26:261–5.
Li R, Zhao SL, Mao T, Xu ZJ, Chen WF. QTL analysis on flag leaf trait in rice. Crops. 2010;3:26–9.
Mccouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H, et al. Report on QTL nomenclature. Rice Gen Newsletter. 1997;14:11–3.
Peng MM, Yang GH, Zhang QQ, An BG, Li YS. QTL Analysis for flag leaf morphological traits in rice (Oryza sativa L.) under different genetic backgrounds. Chin J Rice Sci. 2007;21:247–52.
Qi J, Qian Q, Bu QY, Li S, Chen Q, Sun JQ, et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008;147:1947–59.
Shen B, Yu WD, Zhu YJ, Fan YY, Zhuang JY. Fine mapping of a major quantitative trait locus, qFLL6.2, controlling flag leaf length and yield traits in rice (Oryza sativa L.). Euphytica. 2012;184:57–64.
Tian FK, Ruan BP, Yan MX, Ye SF, Peng YL, Dong GJ, et al. Genetic analysis and QTL mapping of mature seed culturability in indica rice. Rice Sci. 2013;20:313–9.
Tong HH, Mei HW, Xing YZ, Cao YP, Yu XQ, Zhang SQ, et al. QTL analysis for morphological and physiological characteristics of flag leaf at the late development stage in rice. Chin J Rice Sci. 2007;21:493–9.
Vandesompele J, De PA, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a twostep SYBR green I real-time RT-PCR. Anal Biochem. 2002;303:95–8.
Wang YP, Zeng JP, Guo LB, Xing YZ, Xu CG, Mei HW, et al. QTL and correlation analysis on characters of top three leaves and panicle weight in rice (Oryza sativa L.). Chin J Rice Sci. 2004;19:13–20.
Wang Y, Cheng LR, Zheng TQ, Sun Y, Zhou Z, Yang J, et al. Response of main effect qtl for plant height and flag leaf width to artificial selection in rice. Chin J Rice Sci. 2009;23:363–70.
Wang P, Zhou GL, Yu HH, Yu SB. Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor Appl Gene. 2011;123:1319–30.
Xiang JJ, Zhang GH, Qian Q, Xue HW. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012;159:1488–500.
Xiao K, Zuo HL, Gong YJ, Zhang JZ, Zhang YJ, Dong YJ. Locating quantitative trait loci affecting flag-leaf shape traits in rice (Oryza sativa L.). J Shanghai Normal Uni (Natural Sciences). 2007;36:66–70.
Xu JJ, Zhao Q, Zhao YF, Zhu L, Xu CW, Gu MH, et al. Mapping of QTLs for flag leaf shape using whole-genome re-sequenced chromosome segment substitution lines in rice. Chin J Rice Sci. 2011;25:483–7.
Yan YM, Wang XX. Genetic studies of the flag leaf morphology of Indica and Japonica hybrid. Hereditas. 1990;12:1–4.
Yan JQ, Zhu J, He CX, Benmoussa M, Wu P. Molecular marker-assisted dissection of genotype × environment interaction for plant type traits in rice (Oryza sativa L.). Crop Sci. 1999;39:538–44.
Yang RC, Yang HJ. Progress of the research on new plant type rice at IRRI. Hybrid Rice. 1998;13:29–31.
Yuan LP. Hybrid rice breeding for super high yield. Hybrid Rice. 1997;12:1–6.
Yue B, Xue WY, Luo LJ, Xing YZ. QTL Analysis for flag leaf characteristics and their relationships with yield and yield traits in rice. Acta Genet Sin. 2006;33:824–32.
Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21:719–35.
Zhou KD, Ma YQ, Liu TQ, Shen MS. The breeding of subspecific heavy ear hybrid rice -exploration about super-high yield breeding of hybrid rice. J Sichuan Agr Univ. 1995;13:403–7.
Zhou LH, Xie YC, Chen T, Zhang YD, Zhu Z, Zhao QY, et al. Relations between flag leaf morphology and yield and QTL analysis of related traits. Jiangsu J Agr Sci. 2012;28:1207–11.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31221004 and 91335105).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declared that they have no competing interests.
Author contributions
Z-YG and QQ designed research; BZ, W-JY, D-YR, Y-LP, YG, B-PR, LW and G-HZ performed research; BZ, W-JY, PT and L-BG analyzed data; BZ and Z-YG wrote the paper. All author read and approved the final manuscript.
Bin Zhang and Weijun Ye contributed equally to this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, B., Ye, W., Ren, D. et al. Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice 8, 2 (2015). https://doi.org/10.1186/s12284-014-0039-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-014-0039-9