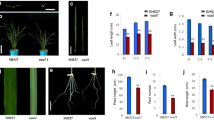
Abstract
Elucidation of the genetic basis of the control of leaf shape could be of use in the manipulation of crop traits, leading to more stable and increased crop production. To improve our understanding of the process controlling leaf shape, we identified a mutant gene in rice that causes a significant decrease in the width of the leaf blade, termed narrow leaf 7 (nal7). This spontaneous mutation of nal7 occurred during the process of developing advanced backcrossed progeny derived from crosses of rice varieties with wild type leaf phenotype. While the mutation resulted in reduced leaf width, no significant morphological changes at the cellular level in leaves were observed, except in bulliform cells. The NAL7 locus encodes a flavin-containing monooxygenase, which displays sequence homology with YUCCA. Inspection of a structural model of NAL7 suggests that the mutation results in an inactive enzyme. The IAA content in the nal7 mutant was altered compared with that of wild type. The nal7 mutant overexpressing NAL7 cDNA exhibited overgrowth and abnormal morphology of the root, which was likely to be due to auxin overproduction. These results indicate that NAL7 is involved in auxin biosynthesis.







Similar content being viewed by others
References
Bartel B (1997) Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 48:51–66
Byrne ME (2005) Networks in leaf development. Curr Opin Plant Biol 8:59–66
Castellano MM, Sablowski R (2005) Intercellular signaling in the transition from stem cells to organogenesis. Curr Opin Plant Biol 8:26–31
Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Gene Dev 20:1790–1799
Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439
Cohen JD, Slovin JP, Hendrickson AM (2003) Two gentically discrete pathways convert tryptophan to auxin: more redundancy in auxin biosysnthesis. Trends Plant Sci 8:197–199
Cui KH, Peng SB, Xing YZ, Yu SB, Xu CG et al (2003) Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor Appl Genet 106:649–658
Fleming AJ (2005) Formation of primordia and phyllotaxy. Curr Opin Plant Biol 8:53–58
Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y et al (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor Appl Genet 108:794–799
Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Genet Genomics 273:150–157
Hake S, Smith HMS, Holtan H, Magnani E, Mele G et al (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20:125–51
Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J et al (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32:573–583
Kepinski S (2006) Integrating hormone signaling and patterning mechanisms in plant development. Curr Opin Plant Biol 9:28–34
Kessler S, Sinha N (2004) Shaping up: the genetic control of leaf shape. Curr Opin Plant Biol 7:65–72
Klee HJ, Horsch RB, Hinchee MA, Hein MB, Hoffmann NL (1987) The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Gene Dev 1:86–96
Li Z, Pinson SRM, Stansel JW, Paterson AH (1998) Genetic dissection of the source–sink relationship affecting fecundity and yield in rice (Oryza sativa L.). Mol Breed 4:419–426
Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28:465–474
Malito E, Alfieri A, Fraaije MW, Mattevi A (2004) Crystal structure of a Baeyer–Villiger monooxygenase. Proc Natl Acad Sci USA 101:13157–13162
Nishimura T, Mori Y, Furukawa T, Kadota A, Koshiba T (2006) Red light causes a reduction in IAA levels at the apical tip by inhibiting de novo biosynthesis from tryptophan in maize coleoptiles. Planta 224:1427–1435
Ozawa K, Kawahigashi H (2006) Positional cloning of the nitrite reductase gene associated with good growth and regeneration ability of calli and establishment of a new selection system for Agrobacterium-mediated transformation in rice (Oryza sativa L.). Plant Sci 170:384–393
Perez-Perez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162:893–915
Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K et al (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426:255–260
Tax FE, Durbak A (2006) Meristems in the movies: live imaging as a tool for decoding intercellular signaling in shoot apical meristems. Plant Cell 18:1331–1337
Tobene-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM et al (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Gene Dev 16:753–763
Tsukaya H (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57:477–496
van Berkel WJH, Kamerbeek NM, Fraaije MW (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124:670–689
Woo YM, Park HJ, Suudi M, Yang JI, Park JJ et al (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65:125–136
Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T et al (2005) Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 139:192–203
Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143:1362–1371
Zazimalova E, Napier RM (2003) Points of regulation for auxin action. Plant Cell Rep 21:625–634
Zhao Y, Christensen SK, Fankhauser C, Cashmen JR, Cohen JD et al (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309
Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J et al (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Gene Dev 16:3100–3112
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Yano.
Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under accession numbers AB354301 and AB354302.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2008_328_MOESM1_ESM.tif
Frequency distribution of the width of the leaf blade in the flag leaf in the advanced backcrossed progeny. Arrowhead indicates the mean of Hayamasari (HY). Three classified genotypes, homozygous for the Italica Livorno allele (black), heterozygous (hatched), and homozygous for the Hayamasari allele (white), assessed using the marker GBR3003, are indicated (TIF 46 kb)
438_2008_328_MOESM2_ESM.tif
Sequence alignment of NAL7. The mutation identified in the nal7 mutant allele is indicated by bold type and by an arrowhead above the aligned sequences. The two conserved motifs are boxed (TIF 72 kb)
438_2008_328_MOESM3_ESM.tif
Phylogenetic tree of the YUCCA gene family. The phylogenetic tree of 14 rice and 11 Arabidopsis YUCCA genes was constructed using CLUSTAL W. Bootstrap analysis values are shown at the nodal branches. The indicated scale represents 0.1 amino acid substitution per site. The accession numbers and plant species are shown in green for Arabidopsis, blue for rice from Yamamoto et al. (2007), and red for rice from this study. Gene ID is according to RAP-DB (http://rapdb.lab.nig.ac.jp/index.html (TIF 62 kb)
438_2008_328_MOESM4_ESM.tif
Expression of nal7 in transgenic plants measured by RT-PCR analysis. Ten independent lines (T1), and Hayamasari and the nal7 mutant as non transgenic plants were used. Total RNA was extracted from the whole shoot of the 6-day-old seedling. The numbers on the right indicate the number of PCR cycles. Ubi2 was used as a loading control (TIF 158 kb)
Rights and permissions
About this article
Cite this article
Fujino, K., Matsuda, Y., Ozawa, K. et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279, 499–507 (2008). https://doi.org/10.1007/s00438-008-0328-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0328-3