Abstract
MicroRNAs (miRNAs) are fundamental post-transcriptional modulators of several critical cellular processes, a number of which are involved in host defense mechanisms. In particular, miRNA let-7 functions as an essential regulator of the function and differentiation of both innate and adaptive immune cells. Let-7 is involved in several human diseases, including cancer and viral infections. Several viral infections have found ways to dysregulate the expression of miRNAs. Extracellular vesicles (EV) are membrane-bound lipid structures released from many types of human cells that can transport proteins, lipids, mRNAs, and miRNAs, including let-7. After their release, EVs are taken up by the recipient cells and their contents released into the cytoplasm. Let-7-loaded EVs have been suggested to affect cellular pathways and biological targets in the recipient cells, and can modulate viral replication, the host antiviral response, and the action of cancer-related viruses. In the present review, we summarize the available knowledge concerning the expression of let-7 family members, functions, target genes, and mechanistic involvement in viral pathogenesis and host defense. This may provide insight into the development of new therapeutic strategies to manage viral infections.
Similar content being viewed by others
Background
MicroRNAs (miRNAs) are small (about 22 nucleotides) RNA molecules that have been shown to regulate gene expression in eukaryotic cells through various mechanisms [1,2,3]. First described in Caenorhabditis elegans, miRNAs have now been found to be widespread in nature. miRNAs have a central role in regulating a number of genes, particularly those genes involved in signaling pathways, and several physiological processes in human cells, including (but not limited to) cellular proliferation, lifespan, metabolism, and cell cycle control [4,5,6]. It is now believed that viruses, which exploit many elements of the host gene expression machinery, are able to encode miRNAs within their genome. Studies over the past decade have elucidated several important roles for viral miRNAs. Furthermore, several host-encoded miRNAs can potentially control viral replication by interacting with target sequences in viral RNAs [6, 7]. miRNAs could have a role as biomarkers of virus-related tumors, and also have some therapeutical potential in cancer treatment. Now, several miRNA-based treatments are being examined in preclinical and clinical trials, for instance, miR-122 in hepatitis C virus (HCV) infection [8]. Besides, virus-mediated changes in miRNA expression can provide an environment that facilitates tumor development [9]. Although some success has been achieved, further studies are still required to fully understand miRNA-based pathways, and virus-related miRNAs.
Extracellular vesicles (EVs) are a family of membrane structures that can be classified based on the vesicle size, function, RNA contents, or biogenesis. According to a classification by the International Society of Extracellular Vesicles, EVs are divided into subclasses including exosomes, microvesicles, and apoptotic bodies [10]. Due to the role of EVs as extracellular transporters of macromolecules, such as proteins and RNA transcripts, they have gained attention for a broad spectrum of applications [10]. EVs can package, release, and transfer miRNAs between cells in a somewhat selective manner. After uptake of the EVs by target cells, the miRNAs are actively released from the EVs. This process protects them against degradation by cell-free RNase enzymes. Viruses employ several mechanisms to evade and suppress the host immune responses, to ensure the establishment and maintenance of viral infections. Nevertheless, the host immune system uses opposing tactics to counteract the viral invasion [11]. Through evolution, viruses have developed the ability to incorporate their nucleic acid components into exosomes, which can exert downstream effects via various mechanisms [12]. Numerous experimental studies have reported the functional transfer of exosomal miRNAs between cells. This transfer facilitates virus replication through suppression of immune responses [6].
The lethal-7 (let-7) gene was first discovered in C. elegans as a key developmental regulator and was one of the first two known microRNAs. In mammals, let-7 is among the miRNAs with the highest expression level in numerous cell types in different species. Increasing evidence has shown the involvement of let-7 family members in critical physiological processes, such as organ development, growth, tissue regeneration, metabolism, and various types of cancer [13]. A vast number of miRNAs have been identified either as tumor inhibitors or as oncogenes (oncomiRs) according to the function of their target genes [4, 13]. Additionally, one individual miRNA can carry out paradoxical dual functions, by acting as a tumor inhibitor in one cancer type while acting as an oncomiR in another cancer type. Let-7 has been reported to possess broad tumor-suppressor effects in a range of cancer types [4]. Besides, many studies have reported the downregulation of let-7 in many viral diseases compared with healthy controls, and thus this miRNA may function as a putative factor encouraging the development of viral infections. Because of the many roles of let-7, modifications of its pathways could play a role in controlling viral infections [e.g., coronavirus disease 2019 (COVID-19), influenza, human immunodeficiency virus (HIV), etc.], as well as cancer-associated viruses. In the present review, we summarize the available knowledge about the expression of the let-7 family, its function, target genes, and mechanisms involved in viral infections, aiming to provide insight into its possible use in the control and therapy of viral infections.
MicroRNA biogenesis and computational resources
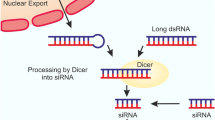
Similar to other RNA transcripts, the biogenesis of miRNAs begins within the cell nucleus. The majority of miRNAs are initially generated as primary transcripts (pri-miRNAs) by RNA polymerase II, and then undergo further processing in the nucleus by RNase III Drosha, which forms long hairpin precursors about 70–100 nucleotides long (termed pre-miRNAs). Thereafter, the pre-miRNAs are exported to the cytoplasm to undergo further maturation. This involves the formation of a complex containing GTP-bound nuclear protein RAN GTPase, the pre-miRNA, and exportin 5 to allow the export of immature miRNAs [14]. After the pre-miRNAs are transported through the nuclear pore complex (NPC), the GTP is hydrolyzed, the NPC is disassembled, and the pre-miRNA is released into the cytosol [15, 16]. In the cytoplasm, the pre-miRNAs are cleaved by RNase III Dicer to form the mature miRNAs. This cleavage eventually leads to a double-stranded 22-nt product, composed of a mature miRNA guide strand and a passenger strand. The miRNA duplex is composed of two complementary strands called 5p and 3p. The passenger strand (annotated *) is typically degraded, whereas the opposite strand (guide strand) binds to the target mRNA sequence. The thermodynamic properties of the duplex seem to determine which strand is selected, because the strand with the weaker binding at the 5′ end of the duplex usually acts as the guide strand. Other essential properties of miRNA guide strands are a U bias at the 5′ end and a high percentage of purines (A/G rich), while the passenger strands possess a C bias at the 5′ end with a pyrimidine-rich (U/C) sequence. Nevertheless, the guide strand could tolerate a single point mutation within the duplex sequence, but this may affect post-transcriptional modification, and the type of proteins associated with Ago2 in the RNA-induced silencing complex (RISC) (e.g., trans-activation response RNA-binding protein versus protein activator of dsRNA-dependent protein kinase) [17,18,19,20]. Thus, both arms of the pre-miRNA hairpin can give rise to biologically functional guide miRNAs [21]. The miRNA guide strand then enters the RISC. Depending on the thermodynamic conditions, occasionally the passenger strand is also loaded into the RISC [22, 23]. The RISC binds to the target mRNA, due to the miRNA interacting with mRNA complementary sequences, resulting in target cleavage and/or translational inhibition (Fig. 1).
MicroRNA biogenesis. miRNAs are produced from miRNA genes, which are transcribed via RNA polymerase II/III to form primary miRNA termed pri-miRNA. Thereafter, pri-miRNA is cleaved by Drosha and DGCR8, and forms precursor microRNA (pre-miRNA), which is exported to the cytoplasm via Ran-GTP and exportin-5 to undergo maturation. miRNA duplex composed of mature miRNA is generated from cleaving the pre-miRNA, which is processed by Dicer and transactivation response element RNA-binding protein (TRBP). The single strand of mature miRNA, containing Ago-2 and GW182 proteins, binds to the complex, namely RISC. This complex modulates miRNA target gene expression by target miRNA cleavage and translation inhibition
miRNAs are generally expressed at low levels, and this expression is different in various tissues and in certain environmental conditions. Considering their relatively small size, identifying miRNAs by their properties remains difficult, and experiments can be expensive, time consuming, and difficult. As a result, computational techniques have been proposed to effectively identify miRNA by their characteristics [24, 25]. Computational techniques take advantage of some features of miRNAs, including their conserved state among different species, the synthesis of stable stem-loop constructions from pre-miRNAs, and the minimum free energy of folding [26, 27]. Different computational approaches have been published in the previous decade. Computational resources containing expression profiles of miRNAs include human miRNA expression database (HMED) and mirEX 2.0 ([28, 29]. HMED and mirEX are considered to be comprehensive analysis programs for identifying datasets of miRNA expression. Moreover, some databases contain differentially expressed (DE) miRs obtained from many tissues; for instance, blood miRs are cell-specific miRNAs obtained from peripheral blood [30]. Likewise, ExCell miRDB is a specialized database containing curated information related to DEmiRs in biofluids [31]. Similarly, miRandola gives detailed information regarding several circulating extracellular miRNAs [32, 33]. In addition, many miRNAs have been identified as critical regulators of gene expression. Moreover, miRNA genes obtained from different species have been reported during the past decade, leading to the extensive expansion of miRBase, which is a core repository of miRNA sequences [34]. The current miRBase release version 21 contains a total of 2588 mature miRNAs and 1881 pre-miRNAs identified in the human genome (GRCh38). Moreover, miRNAs have also been identified in viruses. In a study by Tushcl et al., the miRNAs encoded by Epstein–Barr virus (EBV) were identified. Following this discovery, different viral miRNAs have been reported in other viruses [35, 36]. Currently, VIRmiRNA is a comprehensive database for experimentally verified viral miRNAs, containing 1308 different viral miRNA sequences [37]. A number of computational resources are focused on specific types of diseases; such as OncomiRDB, miRCancer, and dbDEMC, which are catalogs of deregulated miRNAs in different types of cancer [38, 39]. PhenomiR is an online database listing dysregulated miRNAs in different diseases and biological pathways [40]. Moreover, AVIRmiR is a subdatabase of VIRmiRNA [37], containing antiviral miRNAs that have been found to be involved in viral infections.
MicroRNAs as biomarkers
The diagnostic potential of miRNAs has become the most important application in medicine. The term biomarker can involve different types of tests, which can provide information on normal or pathological processes [41]. Biomarkers are becoming more accurate and reliable as tremendous technological progress has recently been achieved. Historically, the first biomarkers were employed by early civilized humans, as simple signs such as pulse rate or urine appearance and odor. Currently, biomarkers often involve specific molecules, mainly proteins or small molecules, that can be found in different types of body fluids using analytical tools [42]. Nevertheless, developing novel protein biomarkers is a costly and time-consuming process that is limited by the lack of clinically significant proteins, structural complexity, and challenges in developing precise analytical methods. To achieve the goals of personalized medicine, novel biomarkers with higher accuracy are required. There are several requirements for developing a good biomarker. First and foremost, it should be readily accessible. In other words, a good biomarker should be detected and measured via minimally invasive procedures. Secondly, it should have the appropriate specificity and sensitivity to be utilized in clinical practice, meaning that the biomarker could be detected prior to the appearance of clinical symptoms, and also its levels would vary according to the clinical stage. Finally, an ideal biomarker should be translatable from the research to the clinical setting [43]. It has been found that free nucleic acids are present in human blood samples [44, 45], and DNA and RNA originating from tumor tissues are constantly released into the plasma of patients who are suffering from cancer [46, 47]. Previously, investigators believed that RNA molecules could not be employed as biomarkers by measurement in blood samples, due to the relatively high levels of nucleases in plasma that potentially degrade nucleic acids [48]. However, with the discovery of miRNAs that were stable in samples of fixed tissue, the idea was revisited [49]. For the first time, in 2008, Lawrie et al. reported that miRNAs could be used as cancer biomarkers. They demonstrated the diagnostic application of miRNAs in patients with diffuse large B-cell lymphoma [50, 51]. Ever since, many studies have been designed to assess the diagnostic ability of miRNAs in a wide variety of human disorders. miRNAs could be superior to conventional biomarkers in many human diseases. Moreover, miRNAs can be easily extracted from human bodily fluids, making them readily accessible. They also have shown satisfactory specificity for specific tissues or cell types and good sensitivity for disease progression state, and many studies have used miRNAs to differentiate different stages of cancer [52] and also for monitoring therapeutic responses [53]. Furthermore, there are already well-established techniques for the detection of DNA and RNA sequences, and the cost and time may be more economical, compared with developing new antibodies for detecting protein biomarkers. Additionally, miRNAs have shown potential for guiding the choice of different possible therapies, more precise diagnosis, and monitoring of the response to treatment. Multimarker panels that can measure several miRNAs at the same time could improve both the diagnosis and prognosis of cancers, as well as many other diseases [54, 55].
Let-7
The lethal-7 (let-7) gene was initially described in C. elegans as a critical gene involved in embryonic development. Let-7 is a highly conserved miRNA across animal species, with 22 nucleotides in length. In C. elegans, the let-7 miRNA family includes nine members: let-7, mir48, mir-84, mir-241, mir-265, mir-793, mir-794, mir-795, and mir-1821, which serve to regulate developmental timing in a sequential order during the larval transition process [13, 56]. The main target genes of let-7 in C. elegans are hbl-1, lin-41, and lin-42 [13]. Worms with mutations in let-7 show developmental timing abnormalities during the larval to adult transition [57, 58]. In Drosophila melanogaster, three miRNAs have been shown to be encoded by the let-7 complex (let-7C): let-7, fly lin-4 (miR-125), and miR100, which show the highest expression in the pupal and adult neuromuscular region. The function of let-7 in Drosophila is primarily to regulate the development transition from the third instar to the pupal stage. Flies with mutations in let-7C showed abnormalities in adult behavior (flight, motility, and fertility), and in juvenile neuromusculature features; however, they appeared normal on external examination. Let-7C has been shown to cause programmed remodeling of the abdominal neuromusculature during larval to adult transition [59]. In mice and humans, the let-7 family has 14 and 13 members, respectively. In humans, these members are let-7a-1, let-7a-2, let-7a3, let-7b, let-7c, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, mir-98, and 202. Among these, let-7a has the most conserved sequence across different animal species from nematode C. elegans to Homo sapiens. In mammals, the expression levels of let-7 are highest during embryogenesis and central nervous system (CNS) development. Nevertheless the let-7 family is not seen in human or mouse embryonic stem cells [13, 60].
Let-7 functions
In most organisms, let-7 expression is increased in the late stages of development, and any alteration in the expression of let-7 can lead to pathological conditions, such as neurodegenerative diseases, cancer, and diabetes [60]. Let-7 is also involved in the regulation of cell signaling pathways. Overexpression of miR-98 inhibited the phosphorylation and repressed the Akt and ERK signaling pathways, which are well known to be implicated in carcinogenesis [61,62,63]. In Ewing’s sarcoma, it was found that let-7 directly downregulated signal transducer and activator of transcription 3 (STAT3), consequently reducing the aggressive phenotype [64]. The STAT3 pathway controls cell cycle and cell survival via regulation of a specific gene set, and so any overactivation can drive cancer progression. Poor outcomes and enhanced resistance to chemotherapy and radiotherapy have been observed following activation of the STAT3 signaling pathway [65, 66]. Let-7 has been reported to activate the WNT signaling pathway via targeting estrogen receptors in breast cancer, and transcription factor 4 (TCF-4; a downstream target of WNT) in hepatocellular carcinoma (HCC), resulting in increased cancer stemness and aggressiveness [67]. The WNT pathway plays a central role in several cellular process, including differentiation, proliferation, and migration, and has been shown to enhance tumor growth and the cancer stem cell phenotype [68, 69]. The data suggests that let-7 prevents the aggressive phenotype via negative regulation of carcinogenic signaling pathways [65].
An anti-let-7 2′-O-Me oligonucleotide increased cancer cell proliferation, highlighting the fact that, in human cells, let-7 exerts its tumor suppressor effects via inhibition of pathways involved in cell proliferation [70]. In another experiment, Johnson et al. demonstrated that let-7 plays a fundamental role in malignancy. The authors reported that let-7 family members acted as tumor suppressors via negative regulation of the let-60/RAS axis [71]. Let-7 inhibited the expression of a set of oncogenes and other genes involved in tumor development and progression. These included RAS, LIN28, PBX3, E2F5, E2F1, Myc, ARID3B, long noncoding RNA (lncRNA) H19, and HMGA2 [61, 63, 72, 73]. Silencing experiments using specific antisense oligonucleotides (ASOs) showed that downregulation of these genes led to tumor suppression, which would normally depend on let-7 [65]. Lan et al. suggested that let-7 acted as a tumor suppressor [74]. They found inhibition of proliferation in HepG2 HCC cells overexpressing let-7g, by repression of the oncogene c-Myc. Gene expression analysis showed a decrease in the corresponding mRNAs and protein levels. Transfection of the cancer cells with a let-7g inhibitor reversed the effects of miRNA overexpression [74]. Let-7g upregulation also enhanced expression of the p16INK4A tumor suppressor protein, indicating that the let-7g tumor suppressor effect is probably due to miRNA direct control of c-Myc in the regulatory axis (c-Myc-Bmi-1-p16) [75, 76]. These data suggest that let-7g may play its role as a tumor inhibitor in HCC through directly repressing the c-Myc oncogene, leading to increased expression of p16INK4A with tumor suppressor effects [74]. Therefore, let-7 is able to suppress the function of several factors involved in oncogenesis.
Paradoxically, let-7 also has shown some pro-oncogenic properties. Although most of the evidence supports the fact that let-7 is a tumor inhibitor in several cancer types, some new studies have suggested let-7 can function as an oncogene. Several studies have shown that the let-7a3 locus is highly methylated in healthy tissue compared with its hypomethylation status in cancer tissues such as lung and ovarian cancer. Moreover, increased expression of mature let-7a has been observed in these cancers [65, 77, 78]. Brueckner et al. showed that let-7a3 overexpression in lung cancer cells caused more aggressive behavior in an anchorage-independent culture experiment. They found alteration in the expression of several factors controlling cell proliferation, and also a number of genes associated with cell adhesion, encouraging tumor progression and metastasis [77]. Also, higher values of several let-7 family members, such as let-7a3, let-7c, and let-7b, were found to be significantly associated with poor prognosis and short overall survival in patients with ovarian and liver cancer [65]. Ma et al. reported that the increased expression of let-7e in esophageal squamous cell carcinoma (ESCC) cells, increased migration and invasion probably through downregulation of the downstream transcription factor ARID3a. ARID3a acts as a negative regulator of pluripotency, so its downregulation may lead to cancer stemness [79, 80]. The let-7 family member mir-98 has been shown to increase chemoresistance in cancer cells via negative regulation of mir-152 mediated by repression of Dicer1. mir-152 and high expression of miR-98 can regulate RAD51 recombinase levels, which were correlated with poor prognosis in patients with epithelial ovarian cancer (EOC) [65, 81]. Overall, these findings highlight the complex relationship between let-7 and cancer cell aggressiveness, underlining the context-dependent role of many miRNAs. It is possible that let-7 in each specific cancer cell targets a set of genes that are particularly expressed in that cell type. Therefore, using an individual miRNA expression profile to develop a “personalized medicine” approach to target let-7 expression in each patient could be necessary.
When any type of infectious agent enters the body, the innate immune system is the first to be activated, which is able to differentiate microbial cells from the host cells [82]. Toll-like receptors (TLRs) play substantial roles in the recognition of invading pathogens and trigger inflammatory responses designed to prime specific adaptive responses to each type of infection. Dysregulation of TLR signaling pathways can promote excessive inflammation, and contribute to the development of diseases, including various types of cancer [83]. Many miRNAs have been identified as key regulators of TLR signaling [84, 85]. Let-7 seems to play a substantial role in the regulation of TLR4 signaling. For instance, after Cryptosporidium parvum infection, let-7i expression was lower, while TLR4 signaling was upregulated [86]. This finding indicates that let-7i may contribute to host immune responses by modulating the C. parvum-induced upregulation of TLR4. In another study, the induction of TLR4 signaling in response to Helicobacter pylori infection was linked with let-7b. TLR4 controls the activation of nuclear factor κB (NF-κB) and the expression of a set of downstream genes involved in inflammation. Let-7 directly targets the TLR4 mRNA to repress translation, and reduces the innate immune response and inflammation after infection [87].
Let-7 also contributes to the function of adaptive immune cells. Let-7 expression affects the differentiation of effector CD8 T cells, which can release effector cytokines and eliminate the infected target cells. Moreover, let-7 affects the differentiation of single positive thymocytes into naive or memory-like CD8+ cytotoxic T lymphocytes (CTLs). In activated CTLs, lowered let-7 levels enhance the clonal expansion and the acquisition of effector functions via negative regulation of its target genes, Myc and eomesodermin (Eomes) [88]. Let-7 can also regulate the promyelocytic leukemia zinc finger (PLZF) transcription factor, which affects the differentiation and effector functions in natural killer T (NKT) cells [89, 90]. Hence, let-7, by repressing PLZF, can control the cellular development of the thymus, activation of B cells, and antibody production [89].
Let-7 and human oncoviruses
Among the approximately 1400 known human pathogens, viruses make up the largest group. Several viruses are carcinogenic, and can induce various cancers in infected patients [91]. Viruses have been implicated in the causation of approximately 14% of cancers, including human papillomavirus (HPV), hepatitis C virus (HCV), hepatitis B virus (HBV), and human herpes virus 8 (HHV8). HBV or HCV are responsible for 80% of HCC cases, the most frequent primary liver malignancy. Some types of HPV, known as high-risk types, are the underlying cause of cervical cancer, as well as several types of head and neck carcinoma. HHV8 and Kaposi’s sarcoma-associated herpesvirus (KSHV) have been identified as causative agents for Kaposi’s sarcoma, which is typically found in HIV-infected patients. HHV8 is also implicated in the pathogenesis of two uncommon B-cell cancers. It has been estimated that HPV is the leading cause of virus-associated cancer worldwide, accounting for about 600,000 new cases every year. On the other hand, HTLV is the least common cause of virus-associated cancer, with 2100 new cases every year. The majority of virus-associated cancers are seen in developing countries, which highlights the crucial need for public health intervention in those regions [92].
Hepatitis B and C viruses
Hepatocellular carcinoma (HCC) is the most common (85%) among primary liver cancers. Globally, HCC is the sixth most common malignancy and the second highest cause of cancer-related death [93]. Chronic HBV infection is the main underlying cause of HCC in Asian countries, while other causes including chronic HCV infection, alcoholic cirrhosis, and non-alcoholic steatohepatitis (NASH) are the major risk factors in Western countries. Other risk factors are excess alcohol consumption and nonalcoholic fatty liver disease [93, 94]. HBV causes cancer by several mechanisms, including virus DNA integration into chromosomes, epigenetic alterations like methylation, oxidative stress, and HBV transcriptional activator HBx protein [95].
HBV DNA and hepatitis B e antigen (HBeAg) can be measured in the blood circulation of HBV-infected patients, and are currently employed in laboratory analysis of affected patients. However, neither can be used as a surrogate marker of viral infection progression and carcinogenesis in clinical settings, because of the heterogeneous nature of HCC. Unfortunately, measurement of the viral load and clinical manifestations are not able to predict the clearance rate and the prognosis in patients with persistent infection. Hence, to optimize the management of HVB-associated diseases, it is important to identify biomarkers and host genetic risk factors, as well as viral and environmental factors. Besides, the occurrence of HCC in patients with cirrhosis is difficult to diagnose, due to the lack of early symptoms [96, 97].
The importance of miRNAs in HCC has been shown by the measurement of miRNA profiles with differential expression in HCC cell lines and tissues compared with normal counterparts [98,99,100,101]. These pioneering studies provided a rationale for examining molecular mechanisms, developing improved diagnostic procedures, and exploring novel therapeutic targets in HCC. High quantities of stable miRNAs have been detected in the circulation in a number of studies, suggesting that differentially expressed miRNAs may serve as reliable fingerprints for many human diseases [4, 5, 102, 103]. For instance, Li et al. [104] examined the hypothesis that the expression profiles of miRNAs could serve as a surrogate marker for the diagnosis of HBV infection, and HBV-positive HCC cases. In this study, 513 patients with HBV (n = 135), HCV (n = 48), and HCC (n = 120) underwent primary screening by the Solexa sequencing procedure, validated by TaqMan probe-based quantitative reverse-transcription PCR (qRT-PCR). The results showed upregulation of both miR-25 and let-7f in plasma samples from patients with HCC; however, no changes were seen in HBV-infected serum samples, and occasionally decreased expression was seen. The authors suggested that some upregulated miRNAs may be involved in HCC development, independent of chronic HBV infection, and could be valuable biomarkers for both HBV infection and HBV-positive HCC patients. Additionally, they reported that miR-375, miR-25, and let-7f showed acceptable receiver operating characteristic (ROC) curves [area under the curve (AUC) of 99.67, sensitivity of 97.9%, and specificity of 99.1%] to distinguish HBV-positive HCC patients from healthy individuals [104].
It was initially believed that HBV could only be transmitted in blood that was positive for the hepatitis B surface antigen (HBsAg); however, it was later shown that the virus could be transmitted by HBsAg-negative blood samples during the seronegative window of the acute infection phase, or in the chronic phase, in the case of occult hepatitis virus B infection (OBI) [105]. HBV DNA can remain in the blood or liver tissue of patients with OBI who were diagnosed as negative for HBsAg. Some patients with OBI may test positive for anti-HBV core antigen- (anti-HBc); therefore, anti-HBc has been employed in screening tests for OBI in blood donors. Nevertheless, it has been shown that > 20% of those who are negative for all virus markers still carry occult HBV infection [106]. Since the main strategy for detection of HBV uses HBsAg, the occult carriers might escape routine screening tests and go on to transmit the virus. Accurate detection of OBI cases is crucial for the elimination of potential viral transmission via blood transfusions [107, 108]. Because there is a possibility that OBI patients are negative for HBsAg and other biomarkers, there is a need for an accurate test for detection of the virus that is equivalent to the HBV DNA PCR assay [109]. Chen et al. [110] used qRT-PCR assays to compare the expression of 13 different HBV-associated miRNAs in serum samples obtained from 11 patients with OBI and 29 healthy subjects. The authors showed that patients with OBI had significantly higher values of miR-23b, miR-150, let-7c, and miR-122 relative to normal subjects. On ROC curve analysis, a signature profile of these four miRNAs could discriminate healthy individuals from OBI patients with an AUC value of 99.9, sensitivity 99.9%, and specificity 99.8% [110] (Table 1).
It has been repeatedly shown that let-7 miRNAs are associated with viral infection, dysfunction of liver cells, and immune response. For example, Shimizu et al. [111] observed that overexpression of let-7 miRNA family members (let-7c or let-7g) led to a significant decrease in anti-apoptotic protein Bcl-xL in two HCC cell lines. This finding suggested that let-7 induced apoptosis via repression of Bcl-xL expression in human HCC. Additionally, OBI can lead to the development of HCC by three possible mechanisms, including production of pro-oncogenic proteins, chronic inflammation leading to hepatic necrosis, and the integration of virus DNA into human liver DNA [112]. Thus, overexpression of let-7 could play a role in the pathogenesis of OBI-related HCC via increasing liver cell damage.
Furthermore, some studies have reported lower values of let-7a in tissues of patients with HCC who are positive for HVB infection compared with healthy controls (Table 1). It has been proposed that let-7a can suppress hepatocyte proliferation by affecting the USP35-ABIN-2 signaling pathway [113], thereby acting as a tumor suppressor in HCC. Recently, Qiu et al. [114] showed that HCC tissue samples expressed let-7a in significantly lower values compared with adjacent normal liver tissue. Moreover, they found higher tissue values of let-7a in patients with highly active HBV replication (HBV DNA > 106 copies/mL) relative to those with less active HBV replication (HBV DNA < 103 copies/mL). In addition, knockdown of let-7a in HepG2.2.15 cells (HepG2 cells engineered to overexpress let-7) using an antisense oligonucleotide resulted in a significant decrease in HBV DNA copy numbers, demonstrating a positive correlation between let-7a and HVB replication. The authors suggested that high levels of let-7a could suppress HCC invasion and proliferation, while it was paradoxically able to enhance HBV replication in hepatocytes [114]. On the other hand, Takata and colleagues [115] reported that mRNA coding for the HBsAg preS2 region in HBV was targeted by the host miRNA let-7g. The expression of HBV mRNAs, such as the preS2 region, could lead to de-repression of the let-7g targets, which could result in long-term oncogenesis. Conversely, let-7g was shown to inhibit the expression of the HBV preS2 protein and other viral proteins. These findings suggested that interactions between the HBV transcripts and the host miRNAs may play a role in the pathogenesis of chronic viral hepatitis [115]. Further studies have reported a negative correlation between the intrahepatic pre-S2 mRNA expression levels and let-7a values [116]. Deng et al. observed that HBV mRNAs repressed let-7a via the miRNA response element, which could lead to universal de-repression of the let-7a host mRNA targets. These changes may contribute to transformation of HCC cells and tumor growth [116]. It is expected that the identification of reciprocal relationships between the viral and host mRNAs may provide insight into HCC pathogenesis, and could inform new therapies against this malignancy.
To date, several studies have shown that the HBV HBx protein can modulate miRNA expression via three main pathways. Firstly, HBx can interact with a transcription factor that regulates miRNA expression [117,118,119]. Secondly, the HBx mRNA can act as a sponge for host miRNAs [120, 121]. Lastly, HBx has been suggested to modulate the biogenesis of pri-miRNA by decreasing the protein levels of Drosha RNase. However, other studies have shown upregulation of several regulators of miRNA biogenesis, such as Drosha, DGCR8, Ago1, and Ago2 [122, 123]. Wang et al. evaluated the expression levels of miRNAs in HBx-expressing cells relative to HepG2 cells (control), using miRNA microarrays. They observed that HBx led to overexpression of 7 miRNAs, while it downregulated 11 other miRNAs, including the let-7 family [124]. HBx-mediated let-7a repression was shown to enhance tumor cell proliferation and promote hepatocarcinogenesis by increasing the expression of transcription factor STAT3. Moreover, it was demonstrated that HBx also downregulated let-7i, which in turn controlled the expression of complement system regulator CD59. Through this mechanism, HBx could protect the HCC cells against complement-dependent cytotoxicity [125]. Overall, HBV proteins have been suggested to contribute to HCC pathogenesis via negative regulation of the let-7 miRNA family members.
HCV accounts for 140,000 new cases of HCC annually [126]. Due to delayed diagnosis, most patients have a poor prognosis; thus, an early diagnosis could improve the survival of many HCC patients [127,128,129,130]. Current HCC guidelines recommend imaging-based diagnosis as the only standard procedure [131, 132]. In this context, much effort has been devoted to find predictive noninvasive biomarkers for the early diagnosis of HCC, and miRNAs are good candidates for this role. Thus, the discovery of predictive biomarkers for the diagnosis and monitoring of HCC is regarded as an urgent issue. Matsuura et al. [133] assessed whether there was any correlation between values of circulating let-7a-5p and the severity of hepatic fibrosis in chronic hepatitis C (CHC) patients. They measured circulating let-7a-5p in serum samples, and in serum-derived extracellular vesicles (EVs) retrieved by a liver biopsy in 84 Japanese patients diagnosed with CHC by qRT-PCR. They investigated the possible correlation between let-7a-5p values and clinicopathological features (histological fibrosis grade, markers of hepatic fibrosis, liver stiffness) in the recruited patients. They found that the circulating levels of let-7a-5p were remarkably lower in patients who were diagnosed with liver cirrhosis. More importantly, transient elastography showed that let-7a-5p serum levels were significantly associated with liver stiffness and hepatic fibrosis markers, including platelet ratio index (APRI), Mac-2 binding protein glycan isomer (M2BPGi), and fibrosis 4 (FIB-4). The ROC curve analysis demonstrated that serum let-7a-5p levels were better for diagnosis of cirrhosis compared with any of the other markers (AUC values of 0.892, 0.800, 0.788, and 0.783 for let-7a-5p, M2BPGi, APRI, and FIB-4, respectively) and were similar to measurement of liver stiffness (AUC 0.909). However, let-7a-5p levels in EVs (AUC 0.681) were lower compared with those in serum. As a result, the authors suggested that circulating let-7a-5p could be a biomarker for predicting the severity of hepatic fibrosis in patients suffering from CHC [133]. Aly et al. measured the expression pattern of the let-7 cluster, including let-7d-1, let7-a-1, and let-7f-1, in serum samples of patients with HCC or chronic HCV infection [134]. The authors found that the serum let-7a-1 levels were remarkably lower in the patients with HCV–HCC compared with the HCV cirrhotic group without HCC. It was also significantly increased in patients with liver cirrhosis compared with the HCV non-cirrhotic group. Additionally, ROC analysis showed that serum let-7a-1 could be a superior biomarker for liver cirrhosis development compared with HCV detection (let-7a-1 AUC 0.768; p = 0.004). They hypothesized that the lower expression of let-7-a1 in serum could promote the development of HCC in chronic HCV patients [134]. However, more studies are needed to assess the clinical application of let-7 detection to diagnose hepatic fibrosis in clinical settings.
miRNA let-7b has been shown to markedly suppress HCV viral replication, and has demonstrated a synergistic effect in combination with the antiviral cytokine interferon-α-2a (IFN-α-2a) in inhibition of HCV infection [135]. Bioinformatics analysis has revealed binding sites for let-7b on the coding region of NS5B and the 5′-untranslated region (UTR) of the HCV genome, which were conserved among different genotypes. Several studies have shown that let-7b is negatively correlated with viral replication and accumulation of HCV RNA, which was not dependent on inhibition of HCV translation. As far as we know, let-7b is the first known miRNA to contain a target site within the coding sequence of the HCV genome.
It was found that let-7b could modulate the expression of IFN-α and IL-28B, and also exert an antiviral effect through repression of HCV protein translation and replication. This was dependent on the host factor, insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1). They also showed that there was a correlation between repression of let-7b and abrogation of the anti-viral effects of IL-28B and IFN-α. Moreover, IL-28B and IFN-α were shown to downregulate IGF2BP1 expression (a target of let-7b), leading to increased antiviral activity of let-7b [136, 137]. Recently, Chen et al. [138] reported that miR-let-7c overexpression significantly repressed the replication of HCV by stimulation of heme oxygenase-1 (HO-1) expression because it inhibited the transcriptional repressor Bach1, eventually resulting in an increased interferon response and suppression of viral protease activity. Accordingly, treatment with a specific inhibitor, exogenous expression of Bach1, and suppression of HO-1 activity and expression all reduced the antiviral activity of miR-let-7c. Taken together, these results highlight the key role of let-7c against HCV infection via targeting Bach1 and consequent transactivation of HO-1-mediated antiviral activity, suggesting a possible role of of let-7c as an antiviral treatment [138].
It has been proposed that let-7a and let-7b can inhibit HCV infection by modulating several cofactors necessary for HCV cell entry, protein production, and RNA replication [139]. There was a negative correlation between the let-7/miR-98 expression levels and the HCV viral load in liver transplantation patients [140]. Upon infection with HCV, some host immune response factors could be modulated by let-7. As previously stated, the let-7b increase following IFN treatment in human hepatocyte HuH7 cells inhibited HCV translation and replication by targeting IGF2BP1 [89, 137]. This implies that let-7b could exert anti-HCV activity via targeting host immune factors and could be used as an anti-HCV therapy and diagnostic test [141]. Recently, Yeh et al. [142] reported that alterations in the expression of let-7b are involved in the progression of HCV infection. Firstly, let-7b can repress the replication of the HCV genome by direct targeting of inhibitors of type 1 IFN signaling. Secondly, let-7b can inhibit the expression of SOCS1 (suppressor of cytokine signaling 1) protein, which acts as a inhibitor of JAK/STAT signaling, leading to increased phosphorylation of STAT1-Y701, thereby increasing the expression of downstream interferon-stimulated genes (ISGs). Let-7 was shown to promote the expression of IFN-β by activation of retinoic acid-inducible gene I (RIG-I) signaling, in addition to direct targeting of the autophagy protein ATG12, and the NF-κB signaling regulator IκB kinase alpha (IKKα) transcripts. This reduced the interaction between RIG-I and the ATG5-ATG12 complex, resulting in increased IFN levels, which in turn activated JAK/STAT signaling. Therefore, the authors concluded that let-7b affected IFN expression by two different signaling pathways [142, 143]. Exploring the regulation of IFN signaling by cellular miRNAs at early stages of HCV infection could clarify the mechanisms underlying primary immune defenses against several types of RNA viruses.
Herpesviruses (EBV and KSHV)
Herpesviruses are a large family of DNA viruses. These viruses can establish lifelong steady-state infections because of their ability to switch between latent (nonproductive) and lytic stages of infection. To date, eight human herpesviruses have been identified, which are classified into three subfamilies: (i) Alphaherpesvirinae including varicella-zoster virus (VZV), human herpes simplex virus type-1 (HSV-1) and type 2 (HSV-2); (ii) Betaherpesvirinae comprising human herpesvirus type 6 (HHV-6) and type 7 (HHV-7), and cytomegalovirus (CMV); (iii) Gammaherpesvirinae including KSHV and EBV [144, 145]. A variety of diseases ranging from cutaneous lesions to several types of cancer can arise following infection with human herpesviruses. The biological life cycle of herpesviruses comprises two major types of replication: latent and lytic replication. The genes that are expressed during the latency period are required for maintenance of the virus genome in an episomal state, while avoiding damage to the host cells. Therefore, this phase allows the viruses to escape the immune responses of infected hosts and establish a persistent infection [146]. Moreover, some coexisting conditions such as immunosuppression allow herpesviruses to switch the life cycle from latent state to lytic infection, leading to viral gene expression and generation of multiple virions [144, 146].
Epstein–Barr virus (EBV) infects the majority of individuals across the world, and has been linked with a number of cancer types, such as gastric carcinoma (GC), B-cell lymphomas (Burkitt and classical Hodgkin), and nasopharyngeal carcinoma (NPC) [147]. The EBV life cycle comprises both latent and lytic infection states in B lymphocytes and epithelial cells. Although EBV remains mostly in the latent phase within B cells, it sometimes switches to the lytic phase to increase cell-to-cell spread. Furthermore, EBV lytic reactivation in the oropharyngeal epithelial cells is required for the generation of infectious viral particles that can be transmitted from host to host. The EBV lytic cycle starts with the expression of the BZLF1 (or Zta) gene, followed by BRLF1 (or Rta) expression. Together these encoded proteins initiate a cascade of subsequent lytic gene expression enabling the biogenesis of linear EBV genomes to be packaged within virions [148]. During the latent phase of infection, a small subset of EBV proteins are expressed and the infected cells become immortalized. The immortalized cells are able to express Epstein–Barr nuclear antigen 1 (EBNA1) during the latent infection [149]. EBNA1 is also the only EBV protein that is crucial for viral replication, and segregation of the viral episomal genomes to maintain a stable copy number of the viral genomes [150, 151]. EBNA1 silencing in GC cell lines was shown to promote EBV reactivation [152]. In 2014, Mansouri et al. [153] observed that several members of let-7 miRNA family (e.g., let-7a) could function as repressors of EBV reactivation in EBV-positive GC and NPC cells. They showed that seven of the let-7 family members were upregulated by EBNA1. EBNA1 overexpression was demonstrated to increase let-7a expression levels in several cell lines, and silencing of EBNA1 reduced let-7a levels. This was proposed to be the mechanism through which EBNA1 could upregulate let-7a expression and inhibit EBV reactivation. Treatment of EBV-expressing cancer cells with a let-7a mimic reduced the percentage of reactivated cells (either via spontaneous reactivation or after chemical treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA) and sodium butyrate), and the use of a let-7 sponge reversed these effects. This finding showed that EBNA1 may enhance latent infection probably via induction of let-7 miRNA. Furthermore, it was found out that Dicer was a downstream target for let-7a and EBNA1, suggesting that high Dicer levels may promote EBV reactivation [153]. The results suggested that host let-7 induced the EBV latent state via negative regulation of Dicer (Fig. 2). The precise mechanism by which increased Dicer levels promote EBV reactivation remains to be explored.
KSHV is a gammaherpesvirus that causes Kaposi’s sarcoma (KS), a malignancy derived from virus-infected endothelial cells in the lining of blood and lymph vessels. KSHV also has been associated with primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD). Similar to other herpesviruses, KSHV can enter a latent stage in the viral life cycle limiting the expression of viral proteins and immunosurveillance responses, thus allowing persistent infection. The virus infects endothelial cells and B lymphocytes in blood and lymph vessels, and shows dual latent and lytic phases [154]. KSHV maintains the latent phase in most infected host cells, and then undergoes reactivation to facilitate cell-to-cell transmission [155]. This dual-phase viral replication is critical for persistent KSHV infection and for the emergence of KS lesions [156]. Among the genes expressed during the latent phase, the latency-associated nuclear antigen (LANA) protein, encoded by the ORF73 (open reading frame 73) is the most characteristic [157]. LANA has been shown to have an important function as a regulator of gene expression and cell proliferation [158]. The switch from latent to lytic phase in KSHV is regulated by the ORF50-encoded replication and transcription activator (RTA) protein [159]. RTA is required to trigger the expression of the cascade of lytic genes in a sequential order for viral reactivation [160]. RTA binds directly to the corresponding response elements located on gene promoters or indirectly by the DNA-binding adaptors (AP-1, C/EBP-a, OCT-1, or RBPJ) [161]. RBPJ (recombination signal binding protein for the immunoglobulin kappa J region) is a well-conserved protein sequence with widespread cellular expression that functions as the main effector in the Notch pathway [162]. RBPJ plays a role in gene silencing via acting as an adaptor of transactivators, and also recruits corepressor complexes [163]. When RTA binds to RBPJ, it causes further self-activation triggering a positive feedback loop that consequently leads to lytic reactivation [164, 165]. The RTA–RBPJ binding can also promote the expression of a number of lytic genes (e.g., ORF50) in addition to the latent phase transcript cluster including LANA [161, 164, 166]. Furthermore, the RTA-induced LANA also binds to RBPJ to form a negative feedback loop repressing RTA expression, resulting in maintenance of the latent virus [166]. Therefore, RBPJ acts as an intermediate in both the positive and negative feedback loops leading to a balanced regulation of viral replication, in two distinct phases of the KSHV life cycle. Although RBPJ–LANA binding leads to downregulation of RTA, RBPJ is required for the lytic phase but not for the latency phase [167]. In the latent phase of infection, RBPJ does not bind to the promoters, but during viral reactivation the same promoters show a high degree of binding of RBPJ after RTA induction [168]. Moreover, latent virus-infected cells show relatively lower expression of RBPJ, suggesting that RBPJ may be regulated by KSHV replication.
In one of the studies on the association between KSHV and Kaposi’s sarcoma, Tan et al. [169] observed in 2015 that the let-7a values in the tumor tissue were significantly lower than in healthy controls. They also found that KSHV reactivation was inhibited by let-7a, by repression of the miRNA target gene, mitogen-activated protein 4 kinase 4 (MAP4K4). The 3′-UTR of RBPJ was also shown to possess a conserved let-7a-binding site, suggesting that let-7a directly targets RBPJ. Recently, Qi et al. [170] demonstrated the role of LANA in repression of KSHV lytic replication through the let-7a/RBPJ axis. LANA was shown to induce let-7a expression along with repression of RBPJ. LANA upregulates the let-7a expression at the transcriptional level by enhancing the cellular notch intracellular domain (NICD) and inducting let-7a maturation via repressing both Lin28B and NF-κB. Let-7a is able to downregulate RBPJ expression via direct interaction with the 3′-UTR of the target mRNA. In this study, silencing of RBPJ resulted in a time- and concentration-dependent repression of KSHV lytic reactivation. Overall, the results suggested that let-7a miRNA overexpression and subsequent repression of RBPJ mediated by LANA, leads to the maintenance of latent viral infection within the host cells [170]. Additionally, the lytic phase of KSHV is characterized by the expression of the ORF50 gene, which serves as an activator of the lytic phase enabling virus replication, virion assembly, and release of the viral progeny. Another KSHV gene, ORF72 encodes a viral homolog of cyclin D, which is not required for transformation of human T lymphocytes [171]. Zhang et al. [172] assessed the effect of let-7 silencing on KSHV lytic replication. The results showed an increase in gene copy number and mRNA transcripts of both ORF50 and ORF72 genes in response to let-7 silencing. Besides, let-7 silencing was shown to increase the protein levels of MAP4K4, COX-2, and phospho-ERK1/2, while the levels of phospho-JNK and phospho-p38 were unchanged. These results demonstrated that silencing of let-7 miRNA could activate KSHV replication, probably through overexpression of MAP4K4 and its downstream mediators, including MMP-13, COX-2, and ERK1/2 phosphorylation, eventually leading to KS development [172].
HPV
Human papillomavirus (HPV) is a member of the family of Papillomaviridae with a broad range of hosts including humans and other animals [173, 174]. Over 200 types of HPV have been identified to cause papillomatosis infections, grouped into high-risk or low-risk strains. To date, 20 high-risk types, including HPV-16, 18, and 45, have been identified as causative agents of several human malignancies, including cervical intraepithelial neoplasia and oropharyngeal and anogenital cancers. It has been proven that HPV-16 and 18 are the major cause of cervical cancer worldwide, accounting for 62.6% and 15.7% of total cases, respectively [175, 176]. The circular genome of HPV encodes six early (E1, E2, E4, E5, E6, E7) and two late (L1, L2) proteins. It has been shown that two oncoproteins, namely, E6 and E7, are directly involved in HPV-induced carcinogenesis [173, 175, 177, 178]. Although high-risk HPV strains are the main cause of cervical cancer, most HPV infections are cleared spontaneously in immunocompetent individuals, and only a minority of HPV-infected women will develop cervical cancer [179]. Therefore, additional factors must contribute to cervical malignant progression.
Recent evidence has implicated STAT3 in cervical carcinogenesis, which shows elevated expression correlated with the disease stage in HPV-16-positive lesions of the cervix [180,181,182]. Increased STAT3 activity has been reported in epithelial transformation, and plays a role in HPV-16-associated cervical carcinogenesis [183]. Thus, the increased activation of the STAT3 pathway is a crucial cellular process linking chronic inflammation to cervical cancer development [184]. However, the cellular mechanisms leading to constitutive activation of the STAT3 pathway and alterations in downstream cellular targets remain largely unclear, and need further study for potential therapeutic targeting. Although several molecules have been identified that can positively or negatively modulate the activity and expression of STAT3, recent studies have highlighted the involvement of miRNAs in governing STAT3 expression and its downstream targets [185]. It has been reported that several miRNAs are aberrantly expressed in HPV-infected cervical cancer cells [186,187,188]. Among the potentially oncogenic miRNAs that could affect the STAT3 pathway, Shishodia and his colleagues investigated the role of two miRNAs, miR-21 and let-7a [189]. The investigation of the miRNA targets showed that let-7a could act as a key regulator of STAT3 expression [124]. Let-7a was found to be often downregulated in cancer cells as a result of chromosomal deletion [190, 191]. Aberrant expression of Let-7a negatively regulates STAT3 transcription via indirect inhibition of IL-6, which is a key cytokine associated with several malignancies, and is also a positive regulator of STAT3 expression [184]. Furthermore, let-7a was observed to directly target STAT3 [124]. Later, Shishodia et al. reported a functional association between the expression levels of miR-21, let-7a, and STAT3 in cervical cancer cells, forming a feedback loop regulated by the oncoprotein E6 [189]. Later, the same researchers [192] measured the levels of miR-21 and let-7a, and investigated their possible correlation with STAT3 in cervical cancer tissues with various grades retrieved from premalignant and malignant lesions in HPV-infected cervical cancer patients. Their results showed that miR-21 was significantly overexpressed. while let-7a was downregulated in cervical cancer tissues. Additionally, miR-21 was directly correlated with the STAT3/ pSTAT3 expression levels, while let-7a demonstrated a reverse correlation in HPV-infected cervical cancer lesions. This reciprocal relationship was not evident for let-7a, especially in precancerous lesions. In HPV-infected lesions, expression levels of miR-21 were correlated with the oncoprotein E6 levels. Contrarily, the let-7a levels were lower in E6-overexpressing lesions, which was consistent with the upregulation of STAT3 mRNA. Unlike miR-21, let-7a showed increased expression in tissue samples from premalignant lesions, compared with higher-grade lesions where let-F7a was downregulated or absent. High expression of let-7a in high-grade squamous intraepithelial lesions (HSIL) but absence in malignant lesions suggests that a major genetic or epigenetic change is involved in malignant switching, which is also required to maintain high levels of STAT3 in malignant cells. More investigations are required to understand the role of let-7a in malignant switching [192]. However, oncoprotein E6 could directly or indirectly via mediators affect the let-7a levels. A negative correlation between let-7a and E6 levels has been found in various clinical samples, while let-7a was also found to inhibit E6 expression in cervical cancer cell lines [189, 192]. Additionally, several studies have shown that silencing of E6 leads to lower STAT3 expression at both transcriptional and translational levels [182, 183, 189]. Moreover, temporary transfection with let-7a mimics (or its precursors with biological function) has been shown to directly and effectively repress STAT3 [189]. Overall, Shishodia et al. hypothesized that in cervical cancer cells there is a strong correlation between lower let-7a expression and increased expression and activation of STAT3, in high-risk HPV-16 infection with higher levels of E6 oncoprotein (Fig. 3). Therefore, miR-21, let-7a, and STAT3 could be used as a potential biomarker signature to discriminate dangerous pre-invasive and malignant cervical lesions, suggesting that STAT3 inhibitors could be tested as a therapy.
Viral respiratory diseases
Respiratory viruses are the most prevalent cause of human infections, with considerable global morbidity and mortality. Respiratory viral infections account for a high economic burden, leading to a large number of people being absent from school and work and many referrals for medical care, and can also exacerbate underlying chronic respiratory diseases, such as chronic obstructive pulmonary disease (COPD) and asthma [193, 194]. The most common viruses causing respiratory infections include orthomyxoviruses, coronaviruses, adenoviruses, paramyxoviruses, picornaviruses, human bocavirus, and human herpesviruses [195]. Although there are some effective vaccines for respiratory viral infections, they may be difficult to access, and there are only a limited number of antiviral medicines for influenza (oseltamivir and zanamivir); however, there are no clinically effective drugs for the most prevalent viral respiratory diseases [196]. Novel antiviral treatments for prophylaxis and therapy of respiratory viral diseases are required according to the World Health Organization (WHO) Battle against Respiratory Viruses (BRaVe) guideline plan [197].
Influenza
Influenza virus (IV) is a member of the Orthomyxoviridae family, which are enveloped viruses with single-stranded, negative-sense, segmented RNA genomes [198]. IVs are divided into three genera, A, B, and C. IV genus A is a common cause of respiratory viral disease in humans and can lead to worldwide pandemics, while IV genus B can causing epidemics (but not pandemics), and IV genus C only causes mild respiratory diseases [199, 200].
A number of cellular miRNAs have been reported to show altered expression during IV infections. miRNAs can control translation of viral mRNAs and also their replication either directly or via mediators, thus affecting viral pathogenesis. From the viewpoint of evolution, it is reasonable to hypothesize that IV employs cellular miRNAs to promote its life cycle because it is an obligate intracellular virus [201]. Let-7 is an important regulator of gene expression in epithelial cells [202], and like the miR-200 family, repression of let-7 causes epithelial features to disappear and fibrotic gene expression is increased [203, 204]. Transient miRNA expression has been shown to be altered after IV infection, as revealed by high-throughput microarray assays and deep sequencing techniques. Studies have reported that the expression levels of let-7 family members, miR-29, and miR-30 are all significantly downregulated post-IV infection [205,206,207]. In another study by Song et al., microarray analysis showed that hsa-let-7g expression was significantly lower in peripheral blood mononuclear cells (PBMCs) obtained from profoundly ill patients diagnosed with H1N1 respiratory infection compared with healthy individuals [208] (Table 1).
Normally, a target mRNA is destroyed when it has a fully matched complementary sequence to the miRNA, while translational repression occurs when the sequences of both RNAs are imperfectly complementary. For instance, Ma et al. [209] reported that several miRNAs showed aberrant expression in IV-positive A549 human lung epithelial cells using high-throughput miRNA microarrays. Among these miRNAs, miR-let-7c in particular showed the highest expression in the lung epithelial cells. Also, they showed that miR-let-7c possessed a fully matching complementary sequence to the 3′-UTR of the H1N1 M1 gene, and could downregulate M1 expression at both the (+) cRNA and protein levels. These results suggested that the let-7c binding to the 3′-UTR of M1 could be used for protection and therapy of IV infections [209].
In a pioneering work, Landgraf et al. produced a comprehensive atlas containing data on the expression of known miRNAs in a large variety of cells and tissues in both mice and humans. They showed that, although most cellular miRNAs are extensively expressed, a number are exclusively expressed in a specific cell type, lineage, or tissue [210]. However, various miRNAs with specific tissue or host expression could affect viral gene expression to different extents. The level of reduction in gene expression is particularly important when aiming to develop a miRNA-attenuated vaccine. IVs used in the production of vaccines provide high titers of virus when grown in chicken egg embryos. To produce IV vaccines in chicken eggs that were attenuated in mammalian cells, Perez et al. [211] evaluated the miRNAs in avian and mammalian cells, and discovered several that were found in mammalian cells but not in eggs. They chose miR-93 to develop an IV vaccine that could replicate in eggs while remaining attenuated in mice [211]. This vaccine demonstrated strong immunity against lethal IV strains in mice. Another potential application of these miRNAs with species-specific attenuation is to increase safety when handling potentially dangerous viruses during molecular research interventions. However, the Perez et al. study had several drawbacks, including that the miR-93 level in lung tissue is rather low, and also the targeted protein in IV is a structural protein and is not involved in viral replication [212]. For this reason, Shen et al. [212] inserted a miRNA recognition element (MRE) for let-7b into the polymerase gene PB1 of IV to develop a modified H1N1 virus with specific replication in the lungs. This was because let-7b is highly expressed in bronchial epithelial cells, and PB1 is an RNA-dependent RNA polymerase that plays a crucial role in viral replication. Insertion of the let-7b-MRE caused the engineered H1N1 to become susceptible to miR-let-7b targeting, showing that viral pathogenicity could be attenuated through incorporation of a replication-restrictive element. Recently, let-7b target sequences were incorporated by Feng et al. into the PB1 of pandemic H1N1 virus identified in 2009 (A/Nanjing/NJU-108/2009) to develop an engineered virus (miRT-H1N1) [213], and the antiviral protection was evaluated after immunization in BALB/c mice. The results showed that the miRT-H1N1 virus was attenuated in infected mice, whereas it maintained wild-type virulence in chicken embryos. Also, the vaccinated mice exhibited strong immunity against lethal A/Nanjing/NJU-108/2009 infection. They suggested that an IV containing an MRE is weakened in vivo and could be used to design a live attenuated vaccine. Overall, miRNAs with specific expression in a certain type of cell or species (e.g., let-7b) could be used to modify pathogenic viral replication to develop live attenuated vaccines. These findings suggest that let-7 miRNA could provide a novel potential biomarker for influenza infection.
SARS CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a member of the family Coronaviridae, subfamily Coronavirinae, genus Betacoronavirus, and was identified as the causative agent of the novel coronavirus disease in 2019 (COVID-19) [214]. This virus was initially identified in a number of pneumonia cases in Wuhan, China in late 2019 [215,216,217]. Now, the pandemic of SARS-CoV-2 has spread around the globe, and has led to the death of a constantly increasing number of people, now over 4.7 million. Over 231 million coronavirus cases have been diagnosed since composing this article [218]. Novel approaches and comprehensively available vaccines are required to control this lethal disease and similar pandemic outbreaks.
Xie et al. [219] hypothesized that let-7 could inhibit COVID-19 by targeting the causative virus. They conducted a series of computational assays to detect putative let-7 target sites located on the SARS-CoV-2 genome, and discovered two sites with complementary sequences matching the seed region of let-7-3p, located in the coding regions of viral S and M proteins. Functional assessment showed that a number of let-7 family members (let-7d, e, f, g, i) and miR-98 could inhibit expression of the S protein, whereas others (let-7b, c, g, i) and miR-98 could inhibit M protein expression. Additionally, some reports showed that let-7a and let-7c were able to inhibit IL-6 expression, which is an inflammatory cytokine highly expressed in COVID-19 patients [184]. They speculated that higher let-7 expression could lower some inflammatory cytokines and chemokines, but not IL-6. This effect could potentially benefit patients by controlling the virus-induced cytokine storm. Xie et al. [219] reported that ectopic expression of let-7a or let-7c decreased IL-6 mRNA, in addition to other SARS-CoV-2-associated inflammatory mediators, such as CCL2, TNF-α, IL-1β, IL-8, VEGFα, and GM-CSF. They also showed that let-7-5p could upregulate IL-6, IL-1β, IL-8, TNF-α, and GM-CSF, while sponging of let-7-3p enhanced GM-CSF, IL-8, TNF-α, and CCL2 expression in THP1 leukemic cell cells that had been treated with lipopolysaccharide (LPS). These findings indicated the potential of let-7 miRNA to suppress damaging inflammatory responses [219].
Approaches to control SARS-CoV-2 infection include targeting the replication machinery of the virus, inhibiting virus binding to host receptors, and blocking the function of viral proteins [220]. It has been reported that cellular miRNAs can inhibit SARS-CoV-2 gene expression at the translational level via binding to 3′-UTR of the targeted viral genes. This can block the host cell receptors and alter viral structural and functional proteins, but does not affect gene expression in the host cells. Demirci et al. identified some potential viral targets that could be affected by human miRNAs. They discovered that several SARS-CoV-2 genes, including ORF6, were affected by multiple host miRNAs. For instance, let-7c-5p was found to target the viral ORF1ab, and inhibit its replication [221]. Sardar et al. identified six human miRNAs that recognized SARS-CoV-2 proteins: let-7a and miR-101 (targeting nonstructural proteins), miR-126 and miR-378 (targeting the N region), and miR-23b (targeting the S region) [222]. Moreover, let-7 could improve immunity against SARS-CoV-2, because the virus suppresses immune responses partly by suppressing host miRNAs.
Respiratory syncytial virus
Respiratory syncytial virus (RSV) is a common pathogenic virus causing pediatric and geriatric infections, with a considerable number of hospitalizations, clinic visits, and > 14, 000 deaths globally each year. RSV belongs to the genus Paramyxovirus with a negative-sense single-stranded RNA (ssRNA) genome, 15 kb in length encoding 11 proteins, that is, NS1, NS2, M, N, P, L, F, G, SH, M2-1, and M2-2. Although, over the past 60 years, scientists have attempted to design an RSV vaccine, these efforts have not yet provided effective agents for prophylaxis and treatment of RSV. These failures are largely due to our lack of knowledge about the host–virus interactions [223]. Some studies have tried to explore the host-to-virus interface to develop a vaccine against RSV. miRNAs have been found to play a role in virus–host interactions, and could be useful in therapeutic and prophylactic strategies. For instance, Bakre et al. [223] reported that RSV-infected-A549 human alveolar epithelial cells showed overexpression of five miRNAs, that is, miR-24, miR-26b, miR-337-3p, let-7f, and miR-520a-5p. They also showed downregulation of two others, miR-595 and miR-198. The virus G protein can modify the expression of some inflammatory factors, whereas it suppressed type I IFN via induction of suppressor of cytokine signaling 1 (SOCS1) and SOCS3 [224, 225]. They infected A549 human alveolar epithelial cells with recombinant RSV (6340WT) or a recombinant virus with mutated G gene (RSVΔG) to investigate the effects of viral G protein on miRNA expression, particularly let-7, and found that RSV G protein increased let-7 expression [223]. They also demonstrated that let-7f controlled the expression of SOCS3 and CCL7/MCP3, which are known to play a role in the host inflammatory response. Previous studies have reported that the G protein inhibited the release of chemokines by bronchoalveolar leukocytes in response to RSV infection [224], but increased the expression of IL-8 [224]. Let-7f was shown to control the expression of early flowering 4 (ELF4), which in turn increases IL-8 expression [226]. The expression of RSV G protein was correlated with let-7f repression, and subsequently reduced the expression of IL-8. It has been reported that some amino acid residues located on the cysteine loop region of RSV G protein contribute to its modulatory effect on IFN-λ expression, and can suppress several miRNAs, including let-7f [227]. Taken together, RSV represses the expression of let-7 miRNA family members to escape from the the host antiviral defense, partly via the crucial RSV G protein.
Let-7 and HIV
The human immunodeficiency virus 1 (HIV-1) is an enveloped RNA virus with a genome composed of two identical single-stranded sequences. HIV-1 is a member of the Lentivirus genus, Retroviridae family, and Orthoretrovirinae subfamily, and is the causative agent of acquired immunodeficiency syndrome (AIDS). HIV-1 initially binds to and replicates inside CD4+ T lymphocytes, as well as monocytes and macrophages. The three stages of HIV-1 infection are the acute phase, chronic phase, and appearance of AIDS. HIV-1 infection may take an average time of 10 years before the development of AIDS [228].
Only low levels of HIV markers (e.g., HIV-1 RNA and p24) can be identified immediately after HIV-1 infection, and during the early viral infection stage these markers may not be detected at all. Therefore, the early stage has been called the “window period” or “eclipse stage” [229]. Typically, HIV-1 markers could be detected 10 days after viral infection, when the viral RNA can be identified using nucleic acid test (NAT) assays [230,231,232]. After that, 15–22 days following infection, HIV-1 p24 antigen becomes detectable. Acute HIV infection is defined as the interval between virus acquisition and development of seroconversion. The acute phase is highly infectious with peak levels of viral load in the circulation, occasionally reaching > 1 × 107copies/mL [233]. Additionally, the viral load accompanied with the absence of neutralizing antibodies produces a high HIV transmission rate [234, 235]. Subsequently, immunoglobulins (IgM and IgG) against HIV-1 appear, which are easily detected using immunoassays [236, 237]. High rates of false-negative results have been reported in the window period of HIV-1 infection [238]. Precise diagnostic approaches for detection of HIV are needed to minimize the risk of HIV transmission from infected individuals. Individuals in the acute phase of HIV infection mostly do not know they have been infected, and may continue to carry out high-risk behavior, thus increasing transmission rates. Accordingly, early screening of HIV-1-infected individuals could help to prevent viral transmission [239]. Therefore, host-associated prognostic and predictive biomarkers are needed in addition to the currently available diagnostic strategies. Identification and validation of cellular miRNAs as potential biomarkers may help us achieve this goal. Recently Biswas et al. [240] assessed the miRNA expression profiles in early-stage HIV-1-infected subjects, in an attempt to create a miRNA-based approach for prediction. They found that four miRNAs showed differential expression in HIV-1-infected individuals (miR-20b-5p, miR-16-5p, miR-223-3p, and miR-195-5p) and could be used to distinguish early infected subjects from non-infected subjects, with high diagnostic power [AUC 1.000 (95% CI 1.00–1.00), sensitivity 100%, and specificity 100%]. In addition, to diagnose the HIV-1-infected individuals within the window period, they created a different four-miRNA-based panel (let-7g-3p, miR-206, miR-16-5p, and miR-181c-3p) also with high diagnostic power [AUC 0.999 (95% CI 0.995–1.000), sensitivity 100%, and specificity 95·8%]. Furthermore, the use of let-7g-3p alone could distinguish early HIV-1-infected subjects from healthy subjects [AUC 0.91 (95% CI 0.81–1.0)] [240] (Table 2).
Interleukin 10 (IL-10) is a multifunctional cytokine that is expressed in a most immune cells [241]. Plasma IL-10 levels have been found to be higher in subjects with HIV-1 infection, and this is thought to contribute to the poor antiviral immune response by CTLs. The levels of IL-10 expression can be controlled at several levels, including the epigenetic level via different signaling pathways, at the transcriptional level via transcription factors, and the post-transcriptional level mediated by miRNAs [242, 243]. Swaminathan et al. [123] showed that let-7 could repress IL-10 expression at the post-transcriptional level. They found that IL-10 was highly upregulated in HUT78 T cells, and proposed that let-7 overexpression decreased IL-10, because silencing of let-7 miRNA led to a significant increase in IL-10 levels. HIV-1 infected HUT78 cells showed lower let-7 levels accompanied by increased IL-10 levels, suggesting that the decreased let-7 level may be involved in the increased IL-10 expression that was seen in HIV-1 infection. Also, they found reduced let-7 levels in primary CD4+ T cells retrieved from blood samples of subjects with HIV-1 infection, compared with non-infected controls, suggesting that the altered miRNA levels could be linked to the increased IL-10 expression in HIV patients. They proposed that dysregulation of the let-7/IL-10 axis could result in the abnormal CTL function seen in HIV-1-infected individuals [123].
Interleukin-2 (IL-2) is an essential cytokine that regulates the cell number, differentiation state, and death of most T cells. IL-2 is mainly released from activated Th cells, and is controlled by several transcription factors, chromatin remodeling agents, and the CD28 co-stimulation pathway [244, 245]. In a number of studies, HIV-1 infection has been shown to suppress IL-2 expression in Th cells in vitro [246,247,248,249]. Recently, Zhang et al. [250] reported that let-7i induced gene expression in Th cells by binding to the TATA-box of the IL-2 promoter, and promoting the assembly of pre-initiation complexes, which are required for mRNA transcription. They observed that HIV-1 infection results in lower levels of mature let-7i, as well as its precursor and primary forms. Additionally, studies have shown that the function of the let-7i promoter is reduced in Th cells following HIV-1 infection. As a result, they suggested that viral infection results in suppression of the let-7i/IL-2 axis contributing to Th cell death. This was a newly described mechanism for HIV-1-induced Th cell death, because IL-2 cytokine can enhance the survival of activated T cells [250]. Furthermore, because IL-2 is known to regulate the balance of the immune system, the let-7i/IL-2 pathway could be responsible for other immune deficiencies seen after HIV-1 infection (e.g., T-cell functional unresponsiveness) [251,252,253].
A number of miRNAs have been proposed to facilitate HIV-1 infection by suppressing proteins involved in the host antiviral response. For instance, p21 protein inhibits HIV-1 gene expression by blockading the CDK9 transcriptional elongation factor, while the TASK1 K+-channel protein inhibits the Vpu-mediated increase in HIV-1 release [254, 255]. Farberov et al. [256] showed that targeted reduction of TASK1 and p21 expression by miR-124a or miR-34a-5p and Let-7c, respectively, could increase HIV-1 replication in HeLa-CCR5 and JLTRG-R5 cells [256].
Extracellular vesicles loaded with let-7 in viral infections
Extracellular vesicles (EV) are membranous structures that have been classified on the basis of vesicle size, function, or biogenesis process. According to the International Society of EVs, they can be divided into microvesicles (MVs), exosomes, and apoptotic bodies (ABs) [10]. Due to the potential capacity of EVs to transport macromolecules, such as proteins and RNA transcripts, from source cells to recipient cells, they have gained considerable attention. Exosome-mediated transfer of mRNAs and miRNAs has been shown to alter processes within the recipient cells, such as regulate protein expression, indicating that exosome-derived RNAs can play functional roles. Moreover the RNA profile of EVs from a daughter cell can vary significantly from the parental cell, which means that cells can change the composition and concentration of RNAs in EVs [11]. Some populations of EVs have been discovered with high amounts of miRNAs amounting to 39% of the entire RNA content of EVs, but only 6% of the total cellular vesicles [257]. This suggested that miRNAs can be selectively and purposefully sorted and packaged into EVs.
The EVs that originate from virus-infected cells have been found to be able to transfer proteins, viral genomes, and host factors from donor to adjacent recipient cells, or from infected tissue to other tissues. This may lead to modulation of the host immune response to encourage the establishment of a productive viral infection [258, 259]. Additionally, the transfer of miRNAs through EVs could facilitate virus spread through modulating immune responses [260, 261]. These finding suggested that exosomes are involved in viral replication; however, the precise mechanisms for this, and the effects on immune system defense, are not yet clarified.
Many studies have evaluated the biological function of different miRNAs in HCV infection. miRNAs within the EVs may enable communication between neighboring cells and affect gene expression in the recipient cells [5]. Matsuura et al. [262] designed a study to assess whether circulating miRNAs contained within EVs could predict disease progression in CHC patients compared with the hepatic expression levels, and investigated the mechanism of the association. They performed a large miRNA profiling study in plasma samples from CHC patients using a microarray technique. The authors found that 323 miRNAs were differentially expressed in CHC patients compared with healthy controls; however, only six of these could distinguish mild hepatitis subjects from patients with severe chronic hepatitis. Let-7d-5p, let-7a, let-7c, and miR-122-5p were promising in terms of predicting disease progression in CHC patients. There was an inverse association between plasma let-7 miRNA and histological fibrosis stage and fibrosis-related markers. Let-7 values in EVs performed similarly compared with plasma values to discriminate hepatic fibrosis. Longitudinal evaluation in a total of CHC 60 patients showed that plasma expression of let-7 significantly decreased over time, in agreement with the progression of liver fibrosis determined by sequential biopsies. They concluded that the let-7 miRNA family showed the best association with the progression of liver fibrosis in CHC patients. Later, the same researchers [133] evaluated the correlation between the serum circulating let-7a-5p values and EVs isolated from the serum of 84 Japanese patients diagnosed with CHC and the correlation with hepatic fibrosis severity in paired liver biopsies. They found that serum let-7a-5p values and let-7a-5p in EVs were significantly reduced in liver cirrhosis patients. Additionally, let-7a-5p values significantly correlated with hepatic fibrosis markers and could predict hepatic cirrhosis more accurately than other markers of hepatic fibrosis [133]. They proposed that the lower levels of let-7 could affect liver fibrogenesis after viral infection by triggering the TGF-β signaling pathway [262].
More than 36 million people around the world are living with HIV infection. Among them, about 2.3 million people are also estimated to be coinfected with HCV [263]. Due to the similar routes of viral transmission and similar high-risk behaviors, HIV-infected patients are at higher risk of HCV infection. Hepatic disorders are now the leading cause of morbidity and death in individuals who are infected with both HIV and HCV. These patients are likely to develop advanced hepatic disease with a more rapid progression rate compared with those who only have HCV mono-infection [264]. Moreover, HCV and HIV exploit the same host exosomal machinery to promote infection and evade the immune response, leading to alterations of the small RNA cargos after viral infection. Martínez-González et al. [263] investigated the small RNA cargo profile of EVs extracted from plasma samples of HIV/HCV-coinfected individuals. They reported that three miRNAs had specific expression in the liver, and miR-21-5p, hsa-miR-let-7a-5p, and hsa-miR-122-5p were upregulated in patients who were coinfected with HIV and HCV, suggesting that EV miRNA cargos could provide information on liver disease progression [263].
Microglia are specific types of macrophages with a mesodermal/mesenchymal origin generally located in the CNS, and are critical mediators of inflammation within the CNS (neuroinflammation) [265]. Neuroinflammation is a characteristic sign of Japanese encephalitis virus (JEV) infection. This virus is neurotropic in nature and typically affects young children (1–5 years old), sometimes resulting in lifelong consequences, like neuronal damage, motor disability, and amnesia [266]. During the past years, it has been found that JEV infection can cause dysregulations in the miRNA profile within the brain, and some miRNAs are involved in the regulation of JEV replication and neuroinflammatory responses [267,268,269]. Nevertheless, the possible role of neurologic cell–cell communication via microglial-derived miRNAs in viral infections has not been explored. Recently, Mukherjee et al. reported that let-7b and let-7a (let-7a/b) miRNAs were upregulated in, and released from, JEV-infected microglial cells incorporated inside EVs [270]. Reportedly, the let -7a/b miRNA could modulate the inflammatory response in microglial cells via activation of the Toll-like receptor 7 (TLR7) signaling pathway [271]; however, the precise role in promoting JEV pathogenesis is unclear. To explore this further, Mukherjee et al. [272] designed a study to clarify the role of let-7a/b in the pathogenesis of JEV. They first evaluated the effect of miRNA-loaded exosomes on primary neurons. They reported that inhibition of TLR7, or addition of let-7a/b, inhibited the JEV-induced activation of the NOTCH pathway, possibly via the NF-κB pathway, and eventually reduced the JEV-induced generation of TNF-α in microglial cells. Additionally, delivery of isolated exosomes released from let-7a/b-overexpressing microglial cells to the brains of healthy mice caused activation of caspases. When neuro2a neuronal cells or primary cortical neurons were incubated with exosomes derived from JEV-infected cells, or with miRNA-overexpressing microglial cells, both induced caspase activation resulting in cell death. Therefore, these findings underline the complex role of let-7a/b involved in JEV pathogenesis. Because let-7a/b can interact with the TLR7 and NOTCH pathways, it can increase TNF-α secretion from microglial cells. Contrarily, exosomes from virus-infected microglia may activate caspases in healthy adjacent neuronal cells, contributing to their death [272] (Table 3).
Conclusion
We have shown that the let-7 family of miRNAs functions as a regulator of a number of crucial cellular processes. We summarized how let-7 miRNAs can affect viral pathogenesis. Investigators are currently attempting to identify new therapeutic drugs for the treatment of viral diseases and cancer-associated viruses. Recently miRNA-based therapeutic approaches have emerged as promising candidates to meet this goal. The role of let-7 role in some viral infections is well understood, whereas its role in other viral diseases (in particular oncovirus infections) is more complicated and still a matter of debate. It has been shown that let-7 miRNAs are able to reduce the expression levels of several target genes, including MAP4K4, STAT3, IL-10, ORF1ab, SARS-CoV-2, and H1N1 M1 genes. These changes in gene expression may affect the antiviral response of the human immune system. There is evidence that let-7 can be extracted from different human bodily fluids, and the expression values of let-7 are different in virus-infected patients relative to healthy individuals, suggesting its potential to be used as a diagnostic biomarker in clinical settings. Moreover, because let-7 plays such a crucial role in the development of virus and cancer-associated virus infections, it could serve as a valuable target for novel treatments of viral infections.
Availability of data and materials
Not applicable.
References
Khani P, Nasri F, Khani-Chamani F, Saeidi F, Sadri-Nahand J, Tabibkhooei A, Mirzaei H. Genetic and epigenetic contribution to astrocytic gliomas pathogenesis. J Neurochem. 2019;148:188–203.
Shabaninejad Z, Yousefi F, Movahedpour A, Ghasemi Y, Dokanehiifard S, Rezaei S, Aryan R, Savardashtaki A, Mirzaei H. Electrochemical-based biosensors for microRNA detection: Nanotechnology comes into view. Analyt Biochem. 2019;581:113349.
Hashemian SM, Pourhanifeh MH, Fadaei S, Velayati AA, Mirzaei H, Hamblin MR. Non-coding RNAs and exosomes: their role in the pathogenesis of sepsis. Mol Ther Nucleic Acids. 2020;21:51–74.
Nahand JS, Karimzadeh MR, Nezamnia M, Fatemipour M, Khatami A, Jamshidi S, Moghoofei M, Taghizadieh M, Hajighadimi S, Shafiee A. The role of miR-146a in viral infection. IUBMB Life. 2020;72:343–60.
Nahand JS, Vandchali NR, Darabi H, Doroudian M, Banafshe HR, Moghoofei M, Babaei F, Salmaninejad A, Mirzaei H. Exosomal microRNAs: novel players in cervical cancer. Epigenomics. 2020;12:1651–60.
Nahand JS, Mahjoubin-Tehran M, Moghoofei M, Pourhanifeh MH, Mirzaei HR, Asemi Z, Khatami A, Bokharaei-Salim F, Mirzaei H, Hamblin MR. Exosomal miRNAs: novel players in viral infection. Epigenomics. 2020;12:353–70.
J.S. Nahand, L. Shojaie, S.A. Akhlagh, M.S. Ebrahimi, H.R. Mirzaei, H.B. Baghi, M. Mahjoubin-Tehran, N. Rezaei, M.R. Hamblin, V. Tajiknia, Cell death pathways and viruses: Role of microRNAs, Molecular Therapy-Nucleic Acids, (2021).
Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–38.
Nahand JS, Rabiei N, Fathazam R, Taghizadieh M, Ebrahimi MS, Mahjoubin-Tehran M, Baghi HB, Khatami A, Abbasi-Kolli M, Mirzaei HR. Oncogenic viruses and chemoresistance: what do we know? Pharmacol Res. 2021;1:105730.
Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:20677.
Groot M, Lee H. Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells. 2020;9:1044.
Urbanelli L, Buratta S, Tancini B, Sagini K, Delo F, Porcellati S, Emiliani C. The role of extracellular vesicles in viral infection and transmission. Vaccines. 2019;7:102.
Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7:100–13.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24.
Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91.
Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6.
Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–76.
Neilsen CT, Goodall GJ, Bracken CP. IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–9.
Ohanian M, Humphreys DT, Anderson E, Preiss T, Fatkin D. A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance. BMC Genet. 2013;14:1–10.
Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans. 2014;42:1135–40.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14.
Yang J-S, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–26.
Yamada Y, Koshizuka K, Hanazawa T, Kikkawa N, Okato A, Idichi T, Arai T, Sugawara S, Katada K, Okamoto Y. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int J Oncol. 2018;52:166–78.
Riolo G, Cantara S, Marzocchi C, Ricci C. miRNA targets: from prediction tools to experimental validation. Methods Protoc. 2021;4:1.
Çakmak E. Computational and Experimental Tools of miRNAs in Cancer. Middle East J Cancer. 2020;11:381–9.
Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm Genome. 2010;21:1–12.
Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: integration of computational and experimental approaches. DNA Cell Biol. 2007;26:321–37.
Gong J, Wu Y, Zhang X, Liao Y, Sibanda VL, Liu W, Guo A-Y. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol. 2014;11:1375–85.
Zielezinski A, Dolata J, Alaba S, Kruszka K, Pacak A, Swida-Barteczka A, Knop K, Stepien A, Bielewicz D, Pietrykowska H. mirEX 2.0—an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015;15:1–9.
Juzenas S, Venkatesh G, Hübenthal M, Hoeppner MP, Du ZG, Paulsen M, Rosenstiel P, Senger P, Hofmann-Apitius M, Keller A. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017;45:9290–301.
Barupal JK, Saini AK, Chand T, Meena A, Beniwal S, Suthar JR, Meena N, Kachhwaha S, Kothari SL. ExcellmiRDB for translational genomics: a curated online resource for extracellular microRNAs. Omics. 2015;19:24–30.
Monga I, Kumar M. Computational resources for prediction and analysis of functional miRNA and their targetome. Methods Mol Biol. 2019;1:215–50.
Karthik L, Kumar G, Keswani T, Bhattacharyya A, Chandar SS, Bhaskara-Rao K. Protease inhibitors from marine actinobacteria as a potential source for antimalarial compound. PLoS ONE. 2014;9:e90972.
Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62.
Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C. Identification of virus-encoded microRNAs. Science. 2004;304:734–6.
K. Plaisance-Bonstaff, R. Renne, Viral miRNAs, Antiviral RNAi, (2011) 43–66.
Qureshi A, Thakur N, Monga I, Thakur A, Kumar M. VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets. Database. 2014;2014:1.
Wang D, Gu J, Wang T, Ding Z. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics. 2014;30:2237–8.
Yang Z, Ren F, Liu C, He S, Sun G, Gao Q, Yao L, Zhang Y, Miao R, Cao Y. dbDEMC: a database of differentially expressed miRNAs in human cancers. BMC Genomics: Springer; 2010. p. 1–8.
Jones SJ, Laskin J, Li YY, Griffith OL, An J, Bilenky M, Butterfield YS, Cezard T, Chuah E, Corbett R. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010;11:1–12.
Pijnappel E, Suurmeijer J, Koerkamp BG, Siveke J, Salvia R, Ghaneh P, Besselink MG, Wilmink J, van Laarhoven H. Mandatory Reporting Measurements in Trials for Potentially Resectable Pancreatic Cancer. Textbook of Pancreatic Cancer: Springer; 2021. p. 107–18.
Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutation Res. 2011;717:85–90.
C.R. Taylor, Introduction to predictive biomarkers: Definitions and characteristics, Predictive Biomarkers in Oncology, Springer. 2019, pp. 3–18.
Bendich A, Wilczok T, Borenfreund E. Circulating DNA as a possible factor in oncogenesis. Science. 1965;148:374–6.
Kamm RC, Smith AG. Nucleic acid concentrations in normal human plasma. Clin Chem. 1972;18:519–22.
Anker P, Lefort F, Vasioukhin V, Lyautey J, Lederrey C, Chen XQ, Stroun M, Mulcahy HE, Farthing M. K-ras mutations are found in DNA extracted from the plasma of patients with colorectal cancer. Gastroenterology. 1997;112:1114–20.
Hasselmann DO, Rappl G, Rössler M, Ugurel S, Tilgen W, Reinhold U. Detection of tumor-associated circulating mRNA in serum, plasma and blood cells from patients with disseminated malignant melanoma. Oncol Rep. 2001;8:115–23.
Kamm R, Smith A. Ribonuclease activity in human plasma. Clin Biochem. 1972;5:198–200.
Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8.
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5.
Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. BioMed Res Int. 2015;2015:1.
Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer—a brief overview. Adv Biol Reg. 2015;57:1–9.
Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM, Voinea SC. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9:276.
Urbanova M, Plzak J, Strnad H, Betka J. Circulating nucleic acids as a new diagnostic tool. Cell Mol Biol Lett. 2010;15:242–59.
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6.
Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69.
Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6.
Sokol NS, Xu P, Jan Y-N, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–6.
Kumar L, Haque R, Nazir A. Role of microRNA Let-7 in modulating multifactorial aspect of neurodegenerative diseases: an overview. Mol Neurobiol. 2016;53:2787–93.
Dai X, Fan W, Wang Y, Huang L, Jiang Y, Shi L, Mckinley D, Tan W, Tan C. Combined delivery of Let-7b microRNA and paclitaxel via biodegradable nanoassemblies for the treatment of KRAS mutant cancer. Mol Pharm. 2016;13:520–33.
Ning Y, Xu M, Cao X, Chen X, Luo X. Inactivation of AKT, ERK and NF-κB by genistein derivative, 7-difluoromethoxyl-5, 4’-di-n-octylygenistein, reduces ovarian carcinoma oncogenicity. Oncol Rep. 2017;38:949–58.
Chen K-J, Hou Y, Wang K, Li J, Xia Y, Yang X-Y, Lv G, Xing X-L, Shen F. Reexpression of Let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. BioMed Res Int. 2014;2014:1.
Zhang Z, Li Y, Huang L, Xiao Q, Chen X, Zhong J, Chen Y, Yang D, Han Z, Shu Y. Let-7a suppresses macrophage infiltrations and malignant phenotype of Ewing sarcoma via STAT3/NF-κB positive regulatory circuit. Cancer Lett. 2016;374:192–201.
Chirshev E, Oberg KC, Ioffe YJ, Unternaehrer JJ. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin Transl Med. 2019;8:1–14.
Bosch-Barrera J, Queralt B, Menendez JA. Targeting STAT3 with silibinin to improve cancer therapeutics. Cancer Treat Rev. 2017;58:61–9.
Sun X, Xu C, Tang S-C, Wang J, Wang H, Wang P, Du N, Qin S, Li G, Xu S. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–9.
Sun H, Ding C, Zhang H, Gao J. Let-7 miRNAs sensitize breast cancer stem cells to radiation-induced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14:3285–92.
Jin B, Wang W, Meng X-X, Du G, Li J, Zhang S-Z, Zhou B-H, Fu Z-H. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:1–10.
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J. The let-7 microRNA represses cell proliferation pathways in human cells. Can Res. 2007;67:7713–22.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47.
Liu Y, Yin B, Zhang C, Zhou L, Fan J. Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma cell lines by targeting c-myc. Biochem Biophys Res Commun. 2012;417:371–5.
Wu A, Wu K, Li J, Mo Y, Lin Y, Wang Y, Shen X, Li S, Li L, Yang Z. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J Transl Med. 2015;13:1–13.
Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, Xie D, He ML, Lin MC, Kung HF. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16INK4A. Int J Cancer. 2011;128:319–31.
Gupta P, Cairns MJ, Saksena NK. Regulation of gene expression by microRNA in HCV infection and HCV–mediated hepatocellular carcinoma. Virol J. 2014;11:1–14.
Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–6.
Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sültmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Can Res. 2007;67:1419–23.
Lu L, Katsaros D, de la Longrais IAR, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Can Res. 2007;67:10117–22.
Ma J, Zhan Y, Xu Z, Li Y, Luo A, Ding F, Cao X, Chen H, Liu Z. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2017;398:37–45.
An G, Miner CA, Nixon JC, Kincade PW, Bryant J, Tucker PW, Webb CF. Loss of Bright/ARID3a function promotes developmental plasticity. Stem Cells. 2010;28:1560–7.
Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X, Li Y, Wu S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9:1–17.
Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–90.
Mukherjee S, Huda S, Sinha-Babu SP. Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand J Immunol. 2019;90:e12771.
Olivieri F, Rippo MR, Prattichizzo F, Babini L, Graciotti L, Recchioni R, Procopio AD. Toll like receptor signaling in “inflammaging”: microRNA as new players. Immun Age. 2013;10:1–10.
He X, Jing Z, Cheng G. MicroRNAs: new regulators of Toll-like receptor signalling pathways. BioMed Res Int. 2014;2014:1.
Chen X-M, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38.
Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. 2015;17:345–56.
Wells AC, Daniels KA, Angelou CC, Fagerberg E, Burnside AS, Markstein M, Alfandari D, Welsh RM, Pobezinskaya EL, Pobezinsky LA. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. eLife. 2017;6:e26398.
Jiang S. Recent findings regarding let-7 in immunity. Cancer Lett. 2018;434:130–1.
Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, Kimura MY, Sharrow SO, Guinter TI, Feigenbaum L. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol. 2015;16:517–24.
Blaylock RL. Viruses and tumor cell microenvironment: a brief summary. Surg Neurol Int. 2019;10:1.
Schiller JT, Lowy DR. Virus infection and human cancer: an overview. Virus Human Cancer. 2014;1:1–10.
Ogunwobi OO, Harricharran T, Huaman J, Galuza A, Odumuwagun O, Tan Y, Ma GX, Nguyen MT. Mechanisms of hepatocellular carcinoma progression. World J Gastroenterol. 2019;25:2279.
Zhang Z, Han Y, Sun G, Liu X, Jia X, Yu X. MicroRNA-325-3p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by down-regulation of aquaporin 5. Cell Mol Biol Lett. 2019;24:1–15.
Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20:11630.
Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, Huang C, Li J, Dong X, Zhou Y. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–77.
Chaiteerakij R, Addissie BD, Roberts LR. Update on biomarkers of hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2015;13:237–45.
Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus–associated hepatocellular carcinoma. Hepatology. 2008;47:1223–32.
Sadri-Nahand J, Bokharaei-Salim F, Salmaninejad A, Nesaei A, Mohajeri F, Moshtzan A, Tabibzadeh A, Karimzadeh M, Moghoofei M, Marjani A. MicroRNAs: key players in virus-associated hepatocellular carcinoma. J Cell Physiol. 2019;234:12188–225.
Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907.
Yousefpouran S, Mostafaei S, Manesh PV, Iranifar E, Bokharaei-Salim F, Nahand JS, Mirzaei H, Taran M, Babaei F, Sayad B. The assessment of selected miRNAs profile in HIV HBV, HCV, HIV/HCV, HIV/HBV co-infection and elite controllers for determination of biomarker. Microb Pathog. 2020;147:104355.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Hussen BM, Ahmadi G, Marzban H, Azar MEF, Sorayyayi S, Karampour R, Nahand JS, Hidayat HJ, Moghoofei M. The role of HPV gene expression and selected cellular miRNAs in lung cancer development. Microb Pathog. 2021;150:104692.
Li L-M, Hu Z-B, Zhou Z-X, Chen X, Liu F-Y, Zhang J-F, Shen H-B, Zhang C-Y, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Can Res. 2010;70:9798–807.
Pollicino T, Raimondo G. Occult hepatitis B infection. J Hepatol. 2014;61:688–9.
Raimondo G, Pollicino T, Romano L, Zanetti A, A. Update on occult hepatitis B infection. Pathol Biol (Paris). 2010;58(2010):254–7.
Allain J-P. Occult hepatitis B virus infection. Transfus Clin Biol. 2004;11:18–25.
Raimondo G, Allain J-P, Brunetto MR, Buendia M-A, Chen D-S, Colombo M, Craxì A, Donato F, Ferrari C, Gaeta GB. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652–7.
Jackson K, Locarnini S, Gish R. Diagnostics of hepatitis B virus: standard of care and investigational. Clin Liver Dis. 2018;12:5.
Chen Y, Li L, Zhou Z, Wang N, Zhang C-Y, Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;201:389–95.
Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H, Doki Y, Mori M, Hayashi N. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704.
Mak L-Y, Wong DK-H, Pollicino T, Raimondo G, Hollinger FB, Yuen M-F. Occult hepatitis B infection and hepatocellular carcinoma: epidemiology, virology, hepatocarcinogenesis and clinical significance. J Hepatol. 2020;73:952–64.
Liu C, Wang L, Chen W, Zhao S, Yin C, Lin Y, Jiang A, Zhang P. USP35 activated by miR let-7a inhibits cell proliferation and NF-κB activation through stabilization of ABIN-2. Oncotarget. 2015;6:27891.
Qiu D, Chen J, Liu J, Luo Z, Jiang W, Huang J, Qiu Z, Yue W, Wu L. Expression of microRNA let-7a positively correlates with hepatitis B virus replication in hepatocellular carcinoma tissues. Exp Biol Med. 2017;242:939–44.
Takata A, Otsuka M, Ohno M, Kishikawa T, Yoshikawa T, Koike K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci Rep. 2016;6:1–10.
Deng M, Hou J, Hu J, Wang S, Chen M, Chen L, Ju Y, Li C, Meng S. Hepatitis B virus mRNAs functionally sequester let-7a and enhance hepatocellular carcinoma. Cancer Lett. 2016;383:62–72.
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:1.
Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–9.
Yip W-K, Cheng AS-L, Zhu R, Lung RW-M, Tsang DP-F, Lau SS-K, Chen Y, Sung JG, Lai PB-S, Ng EK-O. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS ONE. 2011;6:e22888.
Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu X-D. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288:18484–93.
Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol. 2013;87:13370–8.
Liu AM, Zhang C, Burchard J, Fan S, Wong K-F, Dai H, Poon RT, Luk JM. Global regulation on microRNA in hepatitis B virus-associated hepatocellular carcinoma. Omics. 2011;15:187–91.
Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Méndez C, Gelgor L. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188:6238–46.
Wang Y, Lu Y, Toh ST, Sung W-K, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66.
Shan C, Zhang S, Cui W, You X, Kong G, Du Y, Qiu L, Ye L, Zhang X. Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis. 2011;32:1190–7.
de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in a worldwide incidence analysis. Lancet Glob Health. 2018;8(2020):e180–90.
Khalili K, Menezes R, Kim TK, Yazdi LK, Jang H-J, Sharma S, Feld J, Sherman M. The effectiveness of ultrasound surveillance for hepatocellular carcinoma in a Canadian centre and determinants of its success. Can J Gastroenterol Hepatol. 2015;29:267–73.
Dimitroulis D, Golabkesh M, Naguib D, Knoop B, Dannenberg L, Helten C, Pöhl M, Jung C, Kelm M, Zeus T. Safety and efficacy in prasugrel-versus ticagrelor-treated patients with ST-elevation myocardial infarction. J Cardiovasc Pharmacol. 2018;72:186–90.
Mak L-Y, Cruz-Ramón V, Chinchilla-López P, Torres HA, LoConte NK, Rice JP, Foxhall LE, Sturgis EM, Merrill JK, Bailey HH. Global epidemiology, prevention, and management of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book. 2018;38:262–79.
Hartjen P, Höchst B, Heim D, Kammer H, Lucke J, Reinholz M, Baier A, Smeets R, Wege H, Borowski P. The NTPase/helicase domain of hepatitis C virus nonstructural protein 3 inhibits protein kinase C independently of its NTPase activity. Cell Mol Biol Lett. 2013;18:447–58.
Kanda T, Lau GK, Wei L, Moriyama M, Yu M-L, Chuang W-L, Ibrahim A, Lesmana CRA, Sollano J, Kumar M. APASL HCV guidelines of virus-eradicated patients by DAA on how to monitor HCC occurrence and HBV reactivation. Hepatol Int. 2019;13:649–61.
Ayuso C, Rimola J, Vilana R, Burrel M, Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C, Barrufet M. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72–81.
K. Matsuura, N. Aizawa, H. Enomoto, S. Nishiguchi, H. Toyoda, T. Kumada, E. Iio, K. Ito, S. Ogawa, M. Isogawa, Circulating let-7 levels in serum correlate with the severity of hepatic fibrosis in chronic hepatitis C, Open forum infectious diseases, Oxford University Press US, 2018, pp. ofy268.
Aly DM, Gohar NA-H, Abd-El-Hady AA, Khairy M, Abdullatif MM. Serum microRNA let-7a-1/let-7d/let-7f and miRNA 143/145 gene expression profiles as potential biomarkers in HCV induced hepatocellular carcinoma. Asian Pacific J Cancer Prev. 2020;21:555.
Cheng J-C, Yeh Y-J, Tseng C-P, Hsu S-D, Chang Y-L, Sakamoto N, Huang H-D. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621–33.
Fan H-X, Tang H. Complex interactions between microRNAs and hepatitis B/C viruses. World J Gastroenterol. 2014;20:13477.
Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, Jin Q, Yang W. High-throughput profiling of alpha interferon-and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707–18.
Chen WC, Wei CK, Lee JC. Micro RNA-let-7c suppresses hepatitis C virus replication by targeting Bach1 for induction of haem oxygenase-1 expression. J Viral Hepatitis. 2019;26:655–65.
Li Q, Lowey B, Sodroski C, Krishnamurthy S, Alao H, Cha H, Chiu S, El-Diwany R, Ghany MG, Liang TJ. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat Commun. 2017;8:1–16.
Sajjad EA, Radkowski M, Perkowska-Ptasińska A, Pacholczyk M, Durlik M, Fedorowicz M, Pietrzak R, Ziarkiewicz-Wróblewska B, Włodarski P, Malejczyk J. Negative correlation between hepatitis C virus (HCV) and let-7 microRNA family in transplanted livers: the role of rs868 single-nucleotide polymorphism. Ann Transplant. 2017;22:638.
Thibault PA, Wilson JA. Targeting miRNAs to treat hepatitis C virus infections and liver pathology: inhibiting the virus and altering the host. Pharmacol Res. 2013;75:48–59.
Yeh Y-J, Tseng C-P, Hsu S-D, Huang H-Y, Lai MM, Huang H-D, Cheng J-C. Dual effects of Let-7b in the early stage of hepatitis C virus infection. J Virol. 2021;95:e01800-01820.
Ylä-Anttila P. Autophagy receptors as viral targets. Cell Mol Biol Lett. 2021;26:1–11.
J. Ongrádi, Herpesviridae, BoD–Books on Demand2016.
Pang M-F, Lin K-W, Peh S-C. The signaling pathways of Epstein–Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell Mol Biol Lett. 2009;14:222–47.
Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao S-J. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Aids-Associated Viral Oncogenesis: Springer; 2007. p. 69–127.
Shannon-Lowe C, Rickinson A. The global landscape of EBV-associated tumors. Front Oncol. 2019;9:713.
S.C. Kenney, J.E. Mertz, Regulation of the latent-lytic switch in Epstein–Barr virus, Seminars in Cancer Biology, Elsevier, 2014, pp. 60–68.
Klein G. Viral latency and transformation: the strategy of Epstein–Barr virus. Cell. 1989;58:5–8.
Frappier L. EBNA1 and host factors in Epstein–Barr virus latent DNA replication. Curr Opin Virol. 2012;2:733–9.
Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812–5.
Sivachandran N, Wang X, Frappier L. Functions of the Epstein–Barr virus EBNA1 protein in viral reactivation and lytic infection. J Virol. 2012;86:6146–58.
Mansouri S, Pan Q, Blencowe BJ, Claycomb JM, Frappier L. Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. J Virol. 2014;88:11166–77.
Gottwein E. Kaposi’s sarcoma-associated herpesvirus microRNAs. Front Microbiol. 2012;3:165.
Sun R, Tan X, Wang X, Wang X, Yang L, Robertson ES, Lan K. Epigenetic landscape of Kaposi’s sarcoma-associated herpesvirus genome in classic Kaposi’s sarcoma tissues. PLoS Pathog. 2017;13:e1006167.
Chang P-C, Kung H-J. SUMO and KSHV replication. Cancers. 2014;6:1905–24.
Shao Y, Sun K, Xu W, Li X-L, Shen H, Sun W-H. Helicobacter pylori infection, gastrin and cyclooxygenase-2 in gastric carcinogenesis. World J Gastroenterol. 2014;20:12860.
Gjyshi O, Roy A, Dutta S, Veettil MV, Dutta D, Chandran B. Activated Nrf2 interacts with Kaposi’s sarcoma-associated herpesvirus latency protein LANA-1 and host protein KAP1 to mediate global lytic gene repression. J Virol. 2015;89:7874–92.
Zhao Q, Liang D, Sun R, Jia B, Xia T, Xiao H, Lan K. Kaposi’s sarcoma-associated herpesvirus-encoded replication and transcription activator impairs innate immunity via ubiquitin-mediated degradation of myeloid differentiation factor 88. J Virol. 2015;89:415–27.
Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–12.
Persson LM, Wilson AC. Wide-scale use of Notch signaling factor CSL/RBP-Jκ in RTA-mediated activation of Kaposi’s sarcoma-associated herpesvirus lytic genes. J Virol. 2010;84:1334–47.
Wang Y-C, Wang S-H, Wei Y-N, Du D-W, Xu H, Gao C-C, Zheng M-H, Xie J, Li J-C, Dong G-Y. Notch-RBP-J signaling is required by bone marrow stromal cells for the treatment of acute graft versus host disease. Stem Cell Res. 2013;11:721–35.
Lan K, Murakami M, Choudhuri T, Kuppers DA, Robertson ES. Intracellular-activated Notch1 can reactivate Kaposi’s sarcoma-associated herpesvirus from latency. Virology. 2006;351:393–403.
Guito J, Lukac DM. KSHV Rta promoter specification and viral reactivation. Front Microbiol. 2012;3:30.
Palmeri D, Carroll KD, Gonzalez-Lopez O, Lukac DM. Kaposi’s sarcoma-associated herpesvirus Rta tetramers make high-affinity interactions with repetitive DNA elements in the Mta promoter to stimulate DNA binding of RBP-Jk/CSL. J Virol. 2011;85:11901–15.
Lu J, Verma SC, Cai Q, Robertson ES. The single RBP-Jκ site within the LANA promoter is crucial for establishing Kaposi’s sarcoma-associated herpesvirus latency during primary infection. J Virol. 2011;85:6148–61.
Scholz BA, Harth-Hertle ML, Malterer G, Haas J, Ellwart J, Schulz TF, Kempkes B. Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines. PLoS Pathog. 2013;9:e1003336.
Carroll KD, Bu W, Palmeri D, Spadavecchia S, Lynch SJ, Marras SA, Tyagi S, Lukac DM. Kaposi’s sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J Virol. 2006;80:9697–709.
Tan X, Gao Y, Nan Y, Zhang J, Di C, Wang X, Lian F, Cao Y, Hu Y, Xu L. Cellular microRNA Let-7a suppresses KSHV replication through targeting MAP4K4 signaling pathways. PLoS ONE. 2015;10:e0132148.
Qi Y, Zheng G, Di C, Zhang J, Wang X, Hong Y, Song Y, Chen R, Yang Y, Yan Y. Latency-associated nuclear antigen inhibits lytic replication of Kaposi’s sarcoma-associated herpesvirus by regulating let-7a/RBPJ signaling. Virology. 2019;531:69–78.
Ye F, Lei X, Gao S-J. Mechanisms of Kaposi’s sarcoma-associated herpesvirus latency and reactivation. Adv Virol. 2011;2011:1.
Zhang J, Tan X, Yuan Z, Li Y, Qi Y, Nan X, Qi M, Gao H, Lian F, Yang L. Let-7 miRNA silencing promotes Kaposi’s sarcoma-associated herpesvirus lytic replication via activating mitogen-activated protein kinase kinase kinase kinase 4 and its downstream factors. Zhonghua Zhong Liu Za Zhi. 2016;38:485–91.
Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–99.
Khodabandehlou N, Mostafaei S, Etemadi A, Ghasemi A, Payandeh M, Hadifar S, Norooznezhad AH, Kazemnejad A, Moghoofei M. Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer. 2019;19:61.
Wang C, Hai Y, Liu X, Liu N, Yao Y, He P, Dai Q. Prediction of high-risk types of human papillomaviruses using statistical model of protein “sequence space.” Comput Math Methods Med. 2015;2015:1.
Lung MS, Mak WM, Murray V. The use of a human papillomavirus 18 promoter for tissue-specific expression in cervical carcinoma cells. Cell Mol Biol Lett. 2011;16:477–92.
Tang S, Ding S, Yu L, Shen H, Wan Y, Wu Y. Effects of HPV16 E6 protein on Daxx-induced apoptosis in C33A cells. Cell Mol Biol Lett. 2020;25:1–11.
Moghoofei M, Keshavarz M, Ghorbani S, Babaei F, Nahand JS, Tavakoli A, Mortazavi HS, Marjani A, Mostafaei S, Monavari SH. Association between human papillomavirus infection and prostate cancer: a global systematic review and meta-analysis. Asia Pac J Clin Oncol. 2019;15:e59–67.
Nobbenhuis MA, Helmerhorst TJ, van den Brule AJ, Rozendaal L, Voorhorst FJ, Bezemer PD, Verheijen RH, Meijer CJ. Cytological regression and clearance of high-risk human papillomavirus in women with an abnormal cervical smear. Lancet. 2001;358:1782–3.
Takemoto S, Ushijima K, Kawano K, Yamaguchi T, Terada A, Fujiyoshi N, Nishio S, Tsuda N, Ijichi M, Kakuma T. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. Br J Cancer. 2009;101:967–72.
Scheurer ME, Dillon LM, Chen Z, Follen M, Adler-Storthz K. Absolute quantitative real-time polymerase chain reaction for the measurement of human papillomavirus E7 mRNA in cervical cytobrush specimens. Infect Agents Cancer. 2007;2:1–9.
Shukla S, Shishodia G, Mahata S, Hedau S, Pandey A, Bhambhani S, Batra S, Basir SF, Das BC, Bharti AC. Aberrant expression and constitutive activation of STAT3 in cervical carcinogenesis: implications in high-risk human papillomavirus infection. Mol Cancer. 2010;9:1–17.
Shukla S, Mahata S, Shishodia G, Pandey A, Tyagi A, Vishnoi K, Basir SF, Das BC, Bharti AC. Functional regulatory role of STAT3 in HPV16-mediated cervical carcinogenesis. PLoS ONE. 2013;8:e67849.
Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706.
G. Kohanbash, H. Okada, MicroRNAs and STAT interplay, Seminars in cancer biology, Elsevier, 2012, pp. 70–75.
Sadri-Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh MH, Bokharaei-Salim F, Mirzaei H. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer. 2020;146:305–20.
Morgan EL, Patterson MR, Ryder EL, Lee SY, Wasson CW, Harper KL, Li Y, Griffin S, Blair GE, Whitehouse A. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 2020;16:e1008624.
Gómez-Gómez Y, Organista-Nava J, Gariglio P. Deregulation of the miRNAs expression in cervical cancer: human papillomavirus implications. BioMed Res Int. 2013;2013:1.
Shishodia G, Verma G, Srivastava Y, Mehrotra R, Das BC, Bharti AC. Deregulation of microRNAs Let-7a and miR-21 mediate aberrant STAT3 signaling during human papillomavirus-induced cervical carcinogenesis: role of E6 oncoprotein. BMC Cancer. 2014;14:1–13.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004.
Jeong HC, Kim EK, Lee JH, Yoo HN, Kim JK. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol Med Rep. 2011;4:383–7.
Shishodia G, Shukla S, Srivastava Y, Masaldan S, Mehta S, Bhambhani S, Sharma S, Mehrotra R, Das BC, Bharti AC. Alterations in microRNAs miR-21 and let-7a correlate with aberrant STAT3 signaling and downstream effects during cervical carcinogenesis. Mol Cancer. 2015;14:1–13.
Moriyama M, Hugentobler WJ, Iwasaki A. Seasonality of respiratory viral infections. Annu Rev Virol. 2020;7:83–101.
Hershenson MB. Rhinovirus-induced exacerbations of asthma and COPD. Scientifica. 2013;2013:405876.
del Mendoza J, Cornejo-Tapia A, Weilg P, Verne E, Nazario-Fuertes R, Ugarte C, del Valle LJ, Pumarola T. Incidence of respiratory viruses in Peruvian children with acute respiratory infections. J Med Virol. 2015;87:917–24.
McKimm-Breschkin JL, Jiang S, Hui DS, Beigel JH, Govorkova EA, Lee N. Prevention and treatment of respiratory viral infections: presentations on antivirals, traditional therapies and host-directed interventions at the 5th ISIRV Antiviral Group conference. Antiviral Res. 2018;149:118–42.
Legand A, Briand S, Shindo N, Brooks WA, De Jong MD, Farrar J, Aguilera X, Hayden FG. Addressing the public health burden of respiratory viruses: the Battle against Respiratory Viruses (BRaVe) Initiative. Futur Virol. 2013;8:953–68.
Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26:D49–53.
Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–21.
Bhatia A, Kast R. How influenza’s neuraminidase promotes virulence and creates localized lung mucosa immunodeficiency. Cell Mol Biol Lett. 2007;12:111–9.
Zhong Z, Gong W-X, Huang K, Sun X-M, Jin M-L. Regulation of influenza virus infection by microRNAs. J Integr Agric. 2019;18:1421–7.
Shell S, Park S-M, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5.
Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52.
Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–9.
Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio. 2011;2:e00198-e111.
Pociask DA, Robinson KM, Chen K, McHugh KJ, Clay ME, Huang GT, Benos PV, Janssen-Heininger YM, Kolls JK, Anathy V. Epigenetic and transcriptomic regulation of lung repair during recovery from influenza infection. Am J Pathol. 2017;187:851–63.
Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol. 2010;84:3023–32.
Song H, Wang Q, Guo Y, Liu S, Song R, Gao X, Dai L, Li B, Zhang D, Cheng J. Microarray analysis of microRNA expression in peripheral blood mononuclear cells of critically ill patients with influenza A (H1N1). BMC Infect Dis. 2013;13:1–15.
Ma YJ, Yang J, Fan XL, Zhao HB, Hu W, Li ZP, Yu GC, Ding XR, Wang JZ, Bo XC. Cellular micro RNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Mol Med. 2012;16:2539–46.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14.
Perez JT, Pham AM, Lorini MH, Chua MA, Steel J. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–6.
Shen X, Sun W, Shi Y, Xing Z, Su X. Altered viral replication and cell responses by inserting microRNA recognition element into PB1 in pandemic influenza A virus (H1N1) 2009. Mediators Inflamm. 2015;2015:976575.
C. Feng, M. Tan, W. Sun, Y. Shi, Z. Xing. Attenuation of the influenza virus by microRNA response element in vivo and protective efficacy against. pandemic H1N1 virus in mice. Int J Infect Dis. 2009;38(2015):146–52.
Khatami F, Saatchi M, Zadeh SST, Aghamir ZS, Shabestari AN, Reis LO, Aghamir SMK. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci Rep. 2020;10:1–12.
Velavan TP, Meyer CG. The COVID-19 epidemic. Tropical Med Int Health. 2020;25:278.
McIntosh K, Hirsch M, Bloom A. Coronavirus disease (COVID-19): Epidemiology, virology, and prevention. Lancet Infect Dis. 2019;1(2020):2019–20.
F. Oroojalian, A. Haghbin, B. Baradaran, N. Hemat, M.-A. Shahbazi, H.B. Baghi, A. Mokhtarzadeh, M.R. Hamblin, Novel insights into the treatment of SARS-CoV-2 infection: an overview of current clinical trials, Int J Biol Macromol, (2020).
Worldometer, Covid-19, (2020).
Xie C, Chen Y, Luo D, Zhuang Z, Jin H, Zhou H, Li X, Lin H, Zheng X, Zhang J. Therapeutic potential of C1632 by inhibition of SARS-CoV-2 replication and viral-induced inflammation through upregulating let-7. Signal Transduct Target Ther. 2021;6:1–3.
Balmeh N, Mahmoudi S, Mohammadi N, Karabedianhajiabadi A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inform Med Unlocked. 2020;20:100407.
Demirci MDS, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369.
R. Sardar, D. Satish, S. Birla, D. Gupta, Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis, BioRxiv, (2020).
Bakre A, Mitchell P, Coleman JK, Jones LP, Saavedra G, Teng M, Tompkins SM, Tripp RA. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J Gen Virol. 2012;93:2346.
Tripp RA, Jones L, Anderson LJ. Respiratory syncytial virus G and/or SH glycoproteins modify CC and CXC chemokine mRNA expression in the BALB/c mouse. J Virol. 2000;74:6227–9.
Oshansky CM, Krunkosky TM, Barber J, Jones LP, Tripp RA. Respiratory syncytial virus proteins modulate suppressors of cytokine signaling 1 and 3 and the type I interferon response to infection by a toll-like receptor pathway. Viral Immunol. 2009;22:147–61.
Hedvat CV, Yao J, Sokolic RA, Nimer SD. Myeloid ELF1-like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J Biol Chem. 2004;279:6395–400.
Bakre AA, Harcourt JL, Haynes LM, Anderson LJ, Tripp RA. The central conserved region (CCR) of respiratory syncytial virus (RSV) G protein modulates host miRNA expression and alters the cellular response to infection. Vaccines. 2017;5:16.
Doyal L. HIV and AIDS: Putting women on the global agenda. New York: Routledge; 2018.
Fiebig EW, Heldebrant CM, Smith RI, Conrad AJ, Delwart EL, Busch MP. Intermittent low-level viremia in very early primary HIV-1 infection. J Acquir Immune Defic Syndr. 2005;39:133–7.
Vermeulen M, Coleman C, Mitchel J, Reddy R, van Drimmelen H, Fickett T, Busch M, Lelie N. Comparison of human immunodeficiency virus assays in window phase and elite controller samples: viral load distribution and implications for transmission risk. Transfusion. 2013;53:2384–98.
Lindback S, Thorstensson R, Karlsson A, von Sydow M, Flamholc L. Diagnosis of primary HIV-1 infection and duration of follow-up after HIV exposure Karolinska Institute Primary HIV Infection Study Group. AIDS. 2000;14:2333–9.
Lee HY, Giorgi EE, Keele BF, Gaschen B, Athreya GS, Salazar-Gonzalez JF, Pham KT, Goepfert PA, Kilby JM, Saag MS. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261:341–60.
Quinn TC. Acute primary HIV infection. JAMA. 1997;278:58–62.
Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh-Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50.
Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, Kiwanuka N, Kigozi G, Kiddugavu M, Lutalo T. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, Rakai, Uganda. J Infect Dis. 2005;191:1403–9.
Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–63.
Owen SM, Yang C, Spira T, Ou C, Pau C, Parekh B, Candal D, Kuehl D, Kennedy M, Rudolph D. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol. 2008;46:1588–95.
Westheimer E, Fu J, Radix A, Giancotti FR, Hall L, Daskalakis DC, Tsoi B, Peters PJ. An HIV-1 RNA test following a reactive fourth-generation antigen/antibody combination assay confirms a high proportion of HIV infections. J Clin Virol. 2014;61:623–4.
Powers KA, Ghani AC, Miller WC, Hoffman IF, Pettifor AE, Kamanga G, Martinson FE, Cohen MS. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet. 2011;378:256–68.
Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307–16.
Saraiva M, Ogarra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–81.
Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci USA. 2009;106:5761–6.
Brockman MA, Kwon DS, Tighe DP, Pavlik DF, Rosato PC, Sela J, Porichis F, Le Gall S, Waring MT, Moss K. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood J Am Soc Hematol. 2009;114:346–56.
Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature. 2019;568:405–9.
Moon B-I, Kim TH, Seoh J-Y. Functional modulation of regulatory T cells by IL-2. PLoS ONE. 2015;10:e0141864.
Younes S-A, Yassine-Diab B, Dumont AR, Boulassel M-R, Grossman Z, Routy J-P, Sékaly R-P. HIV-1 viremia prevents the establishment of interleukin 2–producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–22.
Sieg SF, Bazdar DA, Harding CV, Lederman MM. Differential expression of interleukin-2 and gamma interferon in human immunodeficiency virus disease. J Virol. 2001;75:9983–5.
Seddiki N, Phetsouphanh C, Swaminathan S, Xu Y, Rao S, Li J, Sutcliffe EL, Denyer G, Finlayson R, Gelgor L. The micro RNA-9/B-lymphocyte-induced maturation protein-1/IL-2 axis is differentially regulated in progressive HIV infection. Eur J Immunol. 2013;43:510–20.
Kassu A, D’Souza M, O’Connor BP, Kelly-McKnight E, Akkina R, Fontenot AP, Palmer BE. Decreased 4-1BB expression on HIV-specific CD4+ T cells is associated with sustained viral replication and reduced IL-2 production. Clin Immunol. 2009;132:234–45.
Zhang Y, Yin Y, Zhang S, Luo H, Zhang H. HIV-1 infection-induced suppression of the Let-7i/IL-2 axis contributes to CD4+ T cell death. Sci Rep. 2016;6:1–11.
McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23.
Yi JS, Cox MA, Zajac AJ. T-cell exhaustion: characteristics, causes and conversion. Immunology. 2010;129:474–81.
Porichis F, Kaufmann DE. HIV-specific CD4 T cells and immune control of viral replication. Curr Opin HIV AIDS. 2011;6:174.
Hsu K, Seharaseyon J, Dong P, Bour S, Marbán E. Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Mol Cell. 2004;14:259–67.
Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121:1549–60.
Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. J Cell Sci. 2015;128:1607–16.
Lee H, Groot M, Pinilla-Vera M, Fredenburgh LE, Jin Y. Identification of miRNA-rich vesicles in bronchoalveolar lavage fluid: Insights into the function and heterogeneity of extracellular vesicles. J Control Release. 2019;294:43–52.
Alenquer M, Amorim M. Exosome biogenesis, regulation, and function in viral infection. Viruses. 2015;7:5066–83.
Liu Z, Zhang X, Yu Q, He JJ. Exosome-associated hepatitis C virus in cell cultures and patient plasma. Biochem Biophys Res Commun. 2014;455:218–22.
Yu X, Odenthal M, Fries J. Exosomes as miRNA carriers: formation–function–future. Int J Mol Sci. 2016;17:2028.
Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinform. 2015;13:17–24.
Matsuura K, De Giorgi V, Schechterly C, Wang RY, Farci P, Tanaka Y, Alter HJ. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology. 2016;64:732–45.
Martínez-González E, Brochado-Kith Ó, Gómez-Sanz A, Martín-Carbonero L, Jimenez-Sousa MÁ, Martínez-Román P, Resino S, Briz V, Fernández-Rodríguez A. Comparison of methods and characterization of small RNAs from plasma extracellular vesicles of HIV/HCV coinfected patients. Sci Rep. 2020;10:1–13.
Medrano LM, Berenguer J, Salgüero S, González-García J, Díez C, Hontañón V, Garcia-Broncano P, Ibañez-Samaniego L, Bellón JM, Jiménez-Sousa MA. Successful HCV therapy reduces liver disease severity and inflammation biomarkers in HIV/HCV-coinfected patients with advanced cirrhosis: a cohort study. Front Med. 2021;8:17.
Graeber MB, Li W, Rodriguez ML. Role of microglia in CNS inflammation. FEBS Lett. 2011;585:3798–805.
Turtle L, Solomon T. Japanese encephalitis—the prospects for new treatments, Nature Reviews. Neurology. 2018;14:298–313.
Goswami S, Banerjee A, Kumari B, Bandopadhyay B, Bhattacharya N, Basu N, Vrati S, Banerjee A. Differential expression and significance of circulating microRNAs in cerebrospinal fluid of acute encephalitis patients infected with Japanese encephalitis virus. Mol Neurobiol. 2017;54:1541–51.
Ashraf U, Zhu B, Ye J, Wan S, Nie Y, Chen Z, Cui M, Wang C, Duan X, Zhang H. MicroRNA-19b-3p modulates Japanese encephalitis virus-mediated inflammation via targeting RNF11. J Virol. 2016;90:4780–95.
Zhu B, Ye J, Nie Y, Ashraf U, Zohaib A, Duan X, Fu ZF, Song Y, Chen H, Cao S. MicroRNA-15b modulates Japanese encephalitis virus–mediated inflammation via targeting RNF125. J Immunol. 2015;195:2251–62.
Mukherjee S, Akbar I, Kumari B, Vrati S, Basu A, Banerjee A. Japanese encephalitis virus-induced let-7a/b interacted with the NOTCH-TLR 7 pathway in microglia and facilitated neuronal death via caspase activation. J Neurochem. 2019;149:518–34.
Massey VL, Qin L, Cabezas J, Caballeria J, Sancho-Bru P, Bataller R, Crews FT. TLR 7-let-7 Signaling contributes to ethanol-induced hepatic inflammatory response in mice and in alcoholic hepatitis. Alcoholism. 2018;42:2107–22.
Mukherjee S, Akbar I, Kumari B, Vrati S, Basu A, Banerjee A. Japanese encephalitis virus-induced let-7a/b interacted with the NOTCH-TLR7 pathway in microglia and facilitated neuronal death via caspase activation. J Neurochem. 2019;149:518–34.
Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, Wang JH, Huang CM, Lee CM, Lin CY, Yen YH. Circulating micro RNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer. 2016;138:714–20.
Orr C, Myers R, Li B, Jiang Z, Flaherty J, Gaggar A, Meissner EG. Longitudinal analysis of serum microRNAs as predictors of cirrhosis regression during treatment of hepatitis B virus infection. Liver Int. 2020;40:1693–700.
X. Hou, Y. Liang, J. Chen, Y. Wei, P. Zeng, L. Wang, C. Lu, H. Diao, Expression profiling of cellular microRNA in asymptomatic HBsAg carriers and chronic hepatitis B patients, BioMed research international, 2017 (2017).
Shi W, Zhang Z, Yang B, Guo H, Jing L, Liu T, Luo Y, Liu H, Li Y, Gao Y. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Medicine. 2017;96:1.
Wakasugi H, Takahashi H, Niinuma T, Kitajima H, Oikawa R, Matsumoto N, Takeba Y, Otsubo T, Takagi M, Ariizumi Y. Dysregulation of miRNA in chronic hepatitis B is associated with hepatocellular carcinoma risk after nucleos (t) ide analogue treatment. Cancer Lett. 2018;434:91–100.
Zhang Z-Z, Liu X, Wang D-Q, Teng M-K, Niu L-W, Huang A-L, Liang Z. Hepatitis B virus and hepatocellular carcinoma at the miRNA level. World J Gastroenterol. 2011;17:3353.
X. Wang, Prospective evidence of a circulating microRNA signature as a non-invasive marker of hepatocellular carcinoma in HBV patients, (2016).
Li J, Shi W, Gao Y, Yang B, Jing X, Shan S, Wang Y, Du Z. Analysis of microRNA expression profiles in human hepatitis B virus-related hepatocellular carcinoma. Clin Lab. 2013;59:1009–15.
Zhu X, Wu L, Xu J, Yang R, Wu F. Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res. 2011;39:2323–9.
Jin H, Lv S, Yang J, Wang X, Hu H, Su C, Zhou C, Li J, Huang Y, Li L. Use of microRNA Let-7 to control the replication specificity of oncolytic adenovirus in hepatocellular carcinoma cells. PLoS ONE. 2011;6:e21307.
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–43.
Wu G, Huang P, Ju X, Li Z, Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med. 2015;8:15108.
Zhang X, Chen C, Wu M, Chen L, Zhang J, Zhang X, Zhang Z, Wu J, Wang J, Chen X. Plasma microRNA profile as a predictor of early virological response to interferon treatment in chronic hepatitis B patients. Antivir Ther. 2012;17:1243.
I. Almas, S. Afzal, M.U. Ashraf, K. Zahid, A. Rasheed, M. Idrees, Studies on circulating microRNAs: Members of Let-7 family and their correlation with Hepatitis C virus disease pathogenesis and treatment concerns, 2017 14th International Bhurban Conference on Applied Sciences and Technology (IBCAST), IEEE, 2017, pp. 174–178.
Chou W-W, Huang C-F, Yeh M-L, Tsai Y-S, Hsieh M-Y, Huang C-I, Huang J-F, Tsai P-C, Hsi E, Juo S-HH. MicroRNA let-7g cooperates with interferon/ribavirin to repress hepatitis C virus replication. J Mol Med. 2016;94:311–20.
Tsai Y-S, Yeh M-L, Tsai P-C, Huang C-I, Huang C-F, Hsieh M-H, Liu T-W, Lin Y-H, Liang P-C, Lin Z-Y. Clusters of circulating let-7 family tumor suppressors are associated with clinical characteristics of chronic hepatitis C. Int J Mol Sci. 2020;21:4945.
Moradifard S, Sharifi Z, Arabkhazaeli A, Ghasemi F. Evaluating serum levels of miR-181a and miR-let7a in HCV infected Iranian patients compared with healthy individuals. Gene Rep. 2021;22:101001.
El-Guendy NM, Helwa R, El-Halawany MS, Ali SAR, Aly MT, Alieldin NH, Fouad SAH, Saeid H, Abdel-Wahab A-HA. The liver microRNA expression profiles associated with chronic hepatitis C virus (HCV) genotype-4 infection: a preliminary study. Hepat Mon. 2016;16:1.
Fawzy IO, Hamza MT, Hosny KA, Esmat G, Abdelaziz AI. Abrogating the interplay between IGF2BP1, 2 and 3 and IGF1R by let-7i arrests hepatocellular carcinoma growth. Growth Factors. 2016;34:42–50.
Gazon H, Belrose G, Terol M, Meniane J-C, Mesnard J-M, Césaire R, Peloponese J-M Jr. Impaired expression of DICER and some microRNAs in HBZ expressing cells from acute adult T-cell leukemia patients. Oncotarget. 2016;7:30258.
Ene AMC, Borze I, Guled M, Costache M, Leen G, Sajin M, Ionica E, Chitu A, Knuutila S. MicroRNA expression profiles in Kaposi’s sarcoma. Pathol Oncol Res. 2014;20:153–9.
O’Hara AJ, Wang L, Dezube BJ, Harrington WJ Jr, Damania B, Dittmer DP. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113:5938–41.
Choi HS, Jain V, Krueger B, Marshall V, Kim CH, Shisler JL, Whitby D, Renne R. Kaposi’s sarcoma-associated herpesvirus (KSHV) induces the oncogenic miR-17–92 cluster and down-regulates TGF-β signaling. PLoS Pathog. 2015;11:e1005255.
Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–45.
Okoye JO, Ngokere AA, Onyenekwe CC, Erinle CA. Comparable expression of miR-let-7b, miR-21, miR-182, miR-145, and p53 in serum and cervical cells: diagnostic implications for early detection of cervical lesions. Int J Health Sci. 2019;13:29.
Barazandeh F, Onsory K, Mohseni-Meybodi A. Association between miR Let-7g gene expression and the risk of cervical cancer in human papillomavirus-infected patients. Feyz J Kashan Univ Med Sci. 2018;21:569–76.
Alles J, Menegatti J, Motsch N, Hart M, Eichner N, Reinhardt R, Meister G, Grässer FA. miRNA expression profiling of Epstein–Barr virus-associated NKTL cell lines by Illumina deep sequencing. FEBS Open Bio. 2016;6:251–63.
Okoye JO, Ngokere AA, Onyenekwe CC, Omotuyi O, Dada DI. Epstein–Barr virus, human papillomavirus and herpes simplex virus 2 co-presence severely dysregulates miRNA expression. Afr J Lab Med. 2021;10:1–10.
Gao L, Ai J, Xie Z, Zhou C, Liu C, Zhang H, Shen K. Dynamic expression of viral and cellular microRNAs in infectious mononucleosis caused by primary Epstein–Barr virus infection in children. Virol J. 2015;12:1–11.
Jonigk D, Izykowski N, Maegel L, Schormann E, Maecker-Kolhoff B, Laenger F, Kreipe H, Hussein K. MicroRNA expression in Epstein–Barr virus-associated post-transplant smooth muscle tumours is related to leiomyomatous phenotype. Clin Sarcoma Res. 2013;3:1–6.
Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. Expression profile of microRNAs in Epstein–Barr virus-infected AGS gastric carcinoma cells. J Virol. 2014;88:1389–93.
Rizzo R, Soffritti I, D’Accolti M, Bortolotti D, Di Luca D, Caselli E. HHV-6A/6B infection of NK cells modulates the expression of miRNAs and transcription factors potentially associated to impaired NK activity. Front Microbiol. 2017;8:2143.
Donyavi T, Bokharaei-Salim F, Baghi HB, Khanaliha K, Janat-Makan MA, Karimi B, Nahand JS, Mirzaei H, Khatami A, Garshasbi S. Acute and post-acute phase of COVID-19: analyzing expression patterns of miRNA-29a-3p, 146a–3p, 155–5p, and let-7b-3p in PBMC. Int Immunopharmacol. 2021;97:107641.
Inchley CS, Sonerud T, Fjærli HO, Nakstad B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect Dis. 2015;15:1–11.
Thornburg NJ, Hayward SL, Crowe JE Jr. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio. 2012;3:e00220-e212.
Makkoch J, Poomipak W, Saengchoowong S, Khongnomnan K, Praianantathavorn K, Jinato T, Poovorawan Y, Payungporn S. Human microRNAs profiling in response to influenza A viruses (subtypes pH1N1, H3N2, and H5N1). Exp Biol Med. 2016;241:409–20.
Zhu Z, Qi Y, Ge A, Zhu Y, Xu K, Ji H, Shi Z, Cui L, Zhou M. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses. 2014;6:1525–39.
Loveday E-K, Svinti V, Diederich S, Pasick J, Jean F. Temporal-and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J Virol. 2012;86:6109–22.
Li Y, Li J, Belisle S, Baskin CR, Tumpey TM, Katze MG, Differential microRNA expression and virulence of avian reassortant, and reconstructed 1918 influenza A viruses. Virology. 1918;421(2011):105–13.
Narla V, Bhakta N, Freedman JE, Tanriverdi K, Maka K, Deeks SG, Ganz P, Hsue P. Unique circulating microRNA profiles in HIV infection. J Acquir Immune Defic Syndr. 2018;79:644–50.
Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:1–15.
Deng J, Ptashkin RN, Wang Q, Liu G, Zhang G, Lee I, Lee YS, Bao X. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Mol Ther Nucleic Acids. 2014;3:e163.
Kumar M, Nerurkar VR. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology. 2014;452:143–51.
Masyeni S, Kuntaman K, Aryati A, Sofro MA, Hadi U, Mastutik G, Purnomo W, Santosa A, Yohan B, Nelwan EJ. Correlation of miR-150, hsa-let-7e, and miR-146a and gene expression of IL-6, IL-8, IP-10, and MIP-1β during dengue virus infection. Narra J. 2021;1:1.
Zhao Z, Zeng J, Luo Z, Dong S, Xie X, Liang X, Yan Y, Liang Q. Functional mapping of AGO-associated zika virus-derived small interfering RNAs in neural stem cells. Front Cell Infect Microbiol. 2021;11:73.
Cowled C, Foo C-H, Deffrasnes C, Rootes CL, Williams DT, Middleton D, Wang L-F, Bean AG, Stewart CR. Circulating microRNA profiles of Hendra virus infection in horses. Sci Rep. 2017;7:1–10.
Engelmann I, Alidjinou EK, Bertin A, Bossu J, Villenet C, Figeac M, Sane F, Hober D. Persistent coxsackievirus B4 infection induces microRNA dysregulation in human pancreatic cells. Cell Mol Life Sci. 2017;74:3851–61.
Zhang S, Gu D, Ouyang X, Xie W. Proinflammatory effects of the hemagglutinin protein of the avian influenza A (H7N9) virus and microRNA-mediated homeostasis response in THP-1 cells. Mol Med Rep. 2015;12:6241–6.
S. Masyeni, U. Hadi, B. Yohan, N. Margyaningsih, R. Sasmono, Detection of micro RNA hsa-let-7e in peripheral blood mononuclear cells infected with dengue virus serotype-2: preliminary study, IOP Conference Series: Earth and Environmental Science, IOP Publishing, 2018, pp. 012010.
Lee Y-R, Yeh S-F, Ruan X-M, Zhang H, Hsu S-D, Huang H-D, Hsieh C-C, Lin Y-S, Yeh T-M, Liu H-S. Honeysuckle aqueous extract and induced let-7a suppress dengue virus type 2 replication and pathogenesis. J Ethnopharmacol. 2017;198:109–21.
Zhou B, Chu M, Xu S, Chen X, Liu Y, Wang Z, Zhang F, Han S, Yin J, Peng B. Hsa-let-7c-5p augments enterovirus 71 replication through viral subversion of cell signaling in rhabdomyosarcoma cells. Cell Biosci. 2017;7:1–14.
Du X, Wang H, Xu F, Huang Y, Liu Z, Liu T. Enterovirus 71 induces apoptosis of SH-SY5Y human neuroblastoma cells through stimulation of endogenous microRNA let-7b expression. Mol Med Rep. 2015;12:953–9.
Lee Y-R, Chang C-M, Yeh Y-C, Huang C-YF, Lin F-M, Huang J-T, Hsieh C-C, Wang J-R, Liu H-S. Honeysuckle aqueous extracts induced let-7a suppress EV71 replication and pathogenesis in vitro and in vivo and is predicted to inhibit SARS-CoV-2. Viruses. 2021;13:308.
Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, Auray G, Gerber M, Fak J, Keller I. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. 2016;19:409–23.
Li N, Du T, Yan Y, Zhang A, Gao J, Hou G, Xiao S, Zhou E-M. MicroRNA let-7f-5p inhibits porcine reproductive and respiratory syndrome virus by targeting MYH9. Sci Rep. 2016;6:1–16.
McGowan K, Simpson KJ, Petrik J. Expression profiles of exosomal microRNAs from HEV-and HCV-infected blood donors and patients: a pilot study. Viruses. 2020;12:833.
Xu TH, Qiu XB, Sheng ZT, Han YR, Wang J, Tian BY, Yao L. Restoration of microRNA-30b expression alleviates vascular calcification through the mTOR signaling pathway and autophagy. J Cell Physiol. 2019;234:14306–18.
Van Ostade X, Dom M, Tjalma W, Van Raemdonck G. Candidate biomarkers in the cervical vaginal fluid for the (self-) diagnosis of cervical precancer. Arch Gynecol Obstet. 2018;297:295–311.
Scheller N, Herold S, Kellner R, Bertrams W, Jung AL, Janga H, Greulich T, Schulte LN, Vogelmeier CF, Lohmeyer J. Proviral microRNAs detected in extracellular vesicles from bronchoalveolar lavage fluid of patients with influenza virus–induced acute respiratory distress syndrome. J Infect Dis. 2019;219:540–3.
Acknowledgements
Not applicable.
Funding
Not relevant.
Author information
Authors and Affiliations
Contributions
HM was involved in the conception, design, statistical analysis, and drafting of the manuscript. AL, SN, MM, AS, SG, MAK, MK, JS, SST, MT, MRH, and NR contributed to data collection and manuscript drafting. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Letafati, A., Najafi, S., Mottahedi, M. et al. MicroRNA let-7 and viral infections: focus on mechanisms of action. Cell Mol Biol Lett 27, 14 (2022). https://doi.org/10.1186/s11658-022-00317-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s11658-022-00317-9