Abstract
Background
Anaplasma phagocytophilum is the etiological agent of granulocytic anaplasmosis in humans and animals. Wild animals and ticks play key roles in the enzootic cycles of the pathogen. Potential ecotypes of A. phagocytophilum have been characterized genetically, but their host range, zoonotic potential and transmission dynamics has only incompletely been resolved.
Methods
The presence of A. phagocytophilum DNA was determined in more than 6000 ixodid ticks collected from the vegetation and wildlife, in 289 tissue samples from wild and domestic animals, and 69 keds collected from deer, originating from various geographic locations in The Netherlands and Belgium. From the qPCR-positive lysates, a fragment of the groEL-gene was amplified and sequenced. Additional groEL sequences from ticks and animals from Europe were obtained from GenBank, and sequences from human cases were obtained through literature searches. Statistical analyses were performed to identify A. phagocytophilum ecotypes, to assess their host range and their zoonotic potential. The population dynamics of A. phagocytophilum ecotypes was investigated using population genetic analyses.
Results
DNA of A. phagocytophilum was present in all stages of questing and feeding Ixodes ricinus, feeding I. hexagonus, I. frontalis, I. trianguliceps, and deer keds, but was absent in questing I. arboricola and Dermacentor reticulatus. DNA of A. phagocytophilum was present in feeding ticks and tissues from many vertebrates, including roe deer, mouflon, red foxes, wild boar, sheep and hedgehogs but was rarely found in rodents and birds and was absent in badgers and lizards. Four geographically dispersed A. phagocytophilum ecotypes were identified, that had significantly different host ranges. All sequences from human cases belonged to only one of these ecotypes. Based on population genetic parameters, the potentially zoonotic ecotype showed significant expansion.
Conclusion
Four ecotypes of A. phagocytophilum with differential enzootic cycles were identified. So far, all human cases clustered in only one of these ecotypes. The zoonotic ecotype has the broadest range of wildlife hosts. The expansion of the zoonotic A. phagocytophilum ecotype indicates a recent increase of the acarological risk of exposure of humans and animals.
Similar content being viewed by others
Background
Anaplasma phagocytophilum is an obligate intracellular bacterium of the family Anaplasmataceae in the order Rickettsiales that causes disease in humans and animals [1]. It infects neutrophils, resulting in influenza-like symptoms clinically and on rare occasions is even a fatal condition in humans [2]. The first cases of Human Granulocytic Anaplasmosis (HGA) were reported from the USA in 1994 [3]. This incidence has increased gradually to 6.1 cases per million persons in 2010. The first European case was reported from Slovenia in 1995. Since, HGA cases have been occasionally reported throughout Europe [4]. It is unclear to what extent HGA poses a risk to public health in Europe: epidemiological data on the disease incidence and disease burden is either incomplete or lacking from most European countries [5]. The non-specificity of the reported symptoms, poor diagnostic tools and lack of awareness of public health professionals further complicate these estimations [4, 6, 7].
Anaplasma phagocytophilum is transmitted to humans by the bite of an infected tick [8]. The main vector in Europe is I. ricinus, which also transmits Borrelia burgdorferi sensu lato, the causative agent of Lyme borreliosis. In the Netherlands, Lyme borreliosis is on the rise: there has been a threefold increase in consultations of general practitioners for tick bites and Lyme borreliosis since 1994. This rise can be partially explained by spatiotemporal increases in the abundance and activity of questing ticks [9–11]. It is to be expected that growth in tick abundance and activity will also increase the risk of human exposure to other tick-borne pathogens such as A. phagocytophilum, but evidence for this extrapolation is lacking [12, 13].
Anaplasma phagocytophilum is maintained in nature through enzootic cycles between ticks and wild animals [8]. The pathogen has been detected in ticks in most European countries and the infection rates range from 0.4% to 67% [4]. In the Netherlands, infection rates in questing nymphs and adults vary between 0% and 8% [12]. Anaplasma phagocytophilum has been detected in a wide range of wildlife species, including ruminants, rodents, insectivores, carnivores, birds and even reptiles [4]. The relative roles of each tick stage and wildlife species in the enzootic life cycles of A. phagocytophilum have not been fully elucidated yet [4].
Anaplasma phagocytophilum is currently considered as a single bacterial species. Cross-infection experiments, where isolates from distinct host origins were not uniformly infectious for heterologous hosts, indicate that host specialization of A. phagocytophilum may occur [14, 15]. Furthermore, Anaplasma phagocytophilum can be genetically divided into either a few or many subclusters, depending on the genetic markers used. As sequence-based clusters in the bacterial world appear to correspond to an “ecotype”, defined as a population of cells in the same ecological niche [16], subclustering may be related to variation in space, host preference, and pathogenicity. Initially, sequences from the 16S rRNA gene have been used for subclustering, but this gene was shown to not be informative enough to delineate distinct ecotypes of A. phagocytophilum[17, 18]. Highly variable gene fragments encoding for major surface proteins [19, 20], and ankA, a secretory protein [21–23], have been used as well. The groEL heat shock operon has an intermediate genetic variability and is expected to act as a marker for demographic analyses [24–27]. Sequences from the groEL operon have been shown to more clearly delineate ecotypes of A. phagocytophilum than do sequences of the 16S rRNA gene [24, 28].
Knowledge on the distribution of A. phagocytophilum in ticks and wildlife in the Netherlands and Belgium is scarce. The aim of this study was to investigate the distribution of A. phagocytophilum in different stages of endemic tick species and in wildlife hosts and free ranging domestic animals. The potential vectors and animal samples were tested by qPCR and conventional PCR, to determine whether they were infected with A. phagocytophilum. We investigated whether genetic delineation, based on groEL, correlates with host distribution/species and zoonotic potential. To assess whether the differential distribution of the genetic variants was due to geographic variation, all available groEL sequences of European A. phagocytophilum isolates were collected and subjected to similar analyses. Population genetic analyses were used to determine which of the ecotypes is expanding.
Methods
Collection of samples and DNA extraction
Questing I. ricinus and Dermacentor reticulatus were collected by blanket dragging at 17 different sites in The Netherlands and Belgium [29, 30]. Ixodes arboricola and I. frontalis (nymphs and adults and fed larvae) were collected from bird nest boxes and from birds that were captured with mistnets and nest traps in forested areas around the city of Antwerp (Belgium) [30]. Ixodes hexagonus feeding on European hedgehogs (Erinaceus europaeus) were collected in a hedgehog-shelter [29]. Ixodes trianguliceps and I. ricinus feeding on bank voles (Myodes glareolus) and wood mice (Apodemus sylvaticus) were collected at several different sites in The Netherlands and Belgium. Ixodes ricinus feeding on red deer (Cervus elaphus), European mouflon (Ovis orientalis musimon), wild boar (Sus scrofa), sheep (Ovis aries), wood mouse (A. sylvaticus), and sand lizard (Lacerta agilis) has been described in previous studies [29, 31, 32]. Ixodes ricinus feeding on roe deer (Capreolus capreolus) were collected at various localities by volunteers from deer-shelters. Deer keds (Lipoptena cervi) were collected from culled roe deer by hunters. Spleen samples were obtained from 19 animal species. These included samples from roe deer, several bird species and badgers (Meles meles), which were found dead or were euthanized and sent to the Dutch Wildlife Health Centre for postmortem examination. The spleen samples obtained from foxes (Vulpes vulpes) as well as the capture of wild rodents have been described elsewhere [29, 33]. EDTA-blood from clinically and laboratory confirmed anaplasmosis from horses were collected in a veterinary hospital [34]. DNA from questing ticks was extracted by alkaline lysis [32]. Blood and spleen samples were kept frozen (−80°C) until testing. DNA from engorged ticks, deer keds, and tissue samples was extracted using the Qiagen DNeasy Blood & Tissue Kit [29].
Polymerase chain reactions and sequencing
All samples were screened for the presence of A. phagocytophilum DNA with a real-time polymerase chain reaction (qPCR) targeting a 77-bp portion of the msp2 gene. The primers used were ApMSP2F (5’-atggaaggtagtgttggttatggtatt-‘3) and ApMSP2R (5’-ttggtcttgaagcgctcgta-‘3), and the probe was ApMSP2P (5’-tggtgccagggttgagcttgagattg-‘3) labeled with FAM6 [35]. This qPCR was performed in a multiplex format with Neoehrlichia mikurensis[29]. qPCR-positive samples were analysed further with primers targeting a fragment of the groEL gene of A. phagocytophilum[36]. All sequences were confirmed by sequencing both strands. The sequences were stored and analysed in Bionumerics (Version 7.1, Applied Math, Belgium), after subtraction of the primer sequences.
Molecular epidemiological database
Anaplasma phagocytophilum groEL DNA sequences with the geographical origin (country) and the host species from which the isolate originated were also downloaded from the Entrez Nucleotide Database (GenBank, NCBI). Anaplasma phagocytophilum sequences originating from Northern white-breasted hedgehogs (Erinaceus roumanicus) were from a previous study [37]. Sequences that did not originate from natural isolates were excluded. Sequences that were too short to cover regions of variation were also excluded from further analysis. A literature search was performed to specifically extract A. phagocytophilum groEL DNA sequences from human patients in Europe [38–43]. DNA sequences and epidemiological data used for this study are given in the Additional file 1: Table S1.
Phylogenetic and population genetic analysis
We delineated four A. phagocytophilum clusters (called ecotypes) by visually inspecting a phylogenetic tree (Additional file 2: Figure S1). A best-scoring maximum likelihood tree was obtained using RAxML 7.5.5 [44] with the option rapid bootstraps (n = 100). Each codon-position was separately analysed using a general-time-reversible model of base substitutions, gamma-distributed rates and invariant proportions. These models of DNA evolution were determined using PartitionFinder 1.0.1 [45]. The sequences were aligned using MAFFT [46] with default options. Alignment was trimmed (position 642 to 1084) to exclude short sequences to visualize genealogy of A. phagocytophilum haplotypes using Haploviewer (http://www.cibiv.at/~greg/haploviewer). Population genetics measures (Ewens-Watterson test, Tajima’s D, Fu’s Fs) were calculated using Arlequin [47] using untrimmed alignment.
Host distributions between and within ecotypes
One A. phagocytophilum ecotype might be over-represented among lysates from a particular host species. We tested this possibility using a multinomial model in which a lysate from a particular host species is evenly associated across all four ecotypes, i.e. with the probability of 1/4 per ecotype. We then estimated by the Monte Carlo method the probability that the number of most numerous ecotypes in a random realization from the multinomial is equal to or greater than the observed maximum among our lysates The probability (i.e. P-values) less than 0.05 were considered significant support for selective distribution. Counting distinct host species is an alternative measure of host diversity per ecotype. However, observed number of distinct host species is best avoided because the sample availability varied by ecotype and a straightforward comparison in this case is invalid. Therefore, we applied the rarefaction analysis to our datasets and calculated whether the differences in observed number of distinct host species per ecotype were statistically significant and not a random variation due to the sampling bias. For this purpose, we computed p-values using EstimateS (Version 9, R. K. Colwell, http://purl.oclc.org/estimates).
Results
A total of 3493 questing nymphs and adult I. ricinus from various geographical areas in the Netherlands and Belgium were tested for the presence of A. phagocytophilum by PCR. DNA of A. phagocytophilum was found in 2.6% of the tested ticks (90/3493), and in 13 of the 17 investigated areas (Table 1). Anaplasma phagocytophilum DNA was also detected in 1.3% (5/386) questing I. ricinus larvae (Table 2). The infection rate of adult I. ricinus was significantly higher than that of larvae or nymphs. No significant difference was observed between the infection rates of larvae and nymphs (Table 2). Anaplasma phagocytophilum DNA was not detected in I. arboricola (n = 79) and I. frontalis (n = 13) collected from nest boxes, nor in questing Dermacentor reticulatus (n = 59), but was found in 42% of the deer keds (29/69) feeding on 10 roe deer.
To investigate the possible vertebrate host species for A. phagocytophilum, tissue samples of many different animals were tested (Table 3). Spleen samples from roe deer (26/38), red foxes (8/81), one wood mouse (1/23) and one common black bird (1/11) were positive (Table 3). Other organs, except brain, of the wood mouse and common black bird were also tested positive in the A. phagocytophilum qPCR (data not shown). Anaplasma phagocytophilum DNA was also amplified from 14 clinically- and laboratory confirmed horses. No A. phagocytophilum DNA was detected in the spleen samples of other rodents (n = 45), insectivores (n = 11), songbirds (n = 26), and badgers (n = 40).
Due to their protected status in the Netherlands and Belgium, it is very difficult to address the infection rate of A. phagocytophilum in wildlife. As a proxy for their infection rates, ticks feeding on wildlife were collected and tested. The infection rates of I. ricinus feeding on roe deer, red deer, hedgehog, sheep, and mouflon (Table 4) were significantly higher than the infection rate of questing adult I. ricinus (Table 2). Ticks from wild boar were also positive (5/48), but not significantly more than I. ricinus from the vegetation (Table 1). Only one of the 109 I. ricinus larvae feeding on wood mice were positive for A. phagocytophilum. This same wood mouse carried 18 A. phagocytophilum- negative larvae (data not shown). Only nine I. trianguliceps feeding on four wood mice (n = 4) and three bank voles (n = 5) were collected. All eight larvae were negative, whereas one female I. trianguliceps feeding on a wood mouse was A. phagocytophilum-positive (Table 4). Both I. ricinus (11/117) and I. frontalis (4/7) feeding on common black birds were A. phagocytophilum-positive (Table 4). One I. frontalis (1/194) and none of the I. arboricola feeding on great/blue tit were A. phagocytophilum-positive. Ixodes ricinus ticks feeding on sand lizards were all negative for A. phagocytophilum (Table 4).
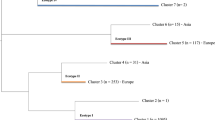
In total, 162 groEL sequences were obtained from the qPCR-positive samples. Together with the groEL sequences from Genbank a phylogenetic tree was reconstructed (Additional file 2: Figure S1). Four major A. phagocytophilum clusters, called ecotypes, could clearly be delineated from this tree. Bootstrap support values were: 98% (ecotype I), 100% (ecotype III) and 100% (ecotype IV). Bootstrap support values for ecotype II was not explicitly computed, but the high support values for the ecotypes I, III and IV imply that the ecotype II is similarly well supported. These four ecotypes were also visually distinguishable in haplotype genealogies in samples both from all over Europe (Figure 1) and from the Netherlands/Belgium (not shown). Based on these four ecotypes, all the available sequences were subdivided further based on their vertebrate host (Table 5). The majority of A. phagocytophilum samples belonged to ecotype I or II (Table 5). Ecotype I was isolated significantly more often from cattle, dogs, hedgehogs, horses, mouflons, red deer, sheep, and humans, while ecotype II was isolated significantly more often from roe deer. Anaplasma phagocytophilum from wood mouse was identical to the groEL sequence found in the engorged I. trianguliceps. Both these samples belonged to ecotype III. Likewise, 18 European isolates from rodents, and two isolates originating from I. persulcatus belonged to ecotype III. Four I. frontalis, one I. ricinus feeding on common blackbirds, and one spleen from a common blackbird contained A. phagocytophilum isolates belonging to ecotype IV. Samples from birds were significantly more often associated with ecotype IV than with other ecotypes (Table 5).
Genealogy of A. phagocytophilum haplotypes. Only 228 isolates of out of 548, representing all 97 haplotypes are shown. Of each haplotype, only one isolate per host from each country is used. An open circle is a haplotype. It is colored by the isolation origin (host species) and drawn in proportion to the sample size. A small blue dot is a missing haplotype. A blue edge is a mutation. The haplotype genealogies were made using Haploviewer software [48]. Roman numerals label the four ecotypes, which were inferred from a phylogenetic tree (Additional file 2: Figure S1).
Ecotype I contained the largest number of distinct hosts, whereas the observed host range of the other three ecotypes was significantly smaller than expected (Table 6), indicating a broad host range for ecotype I and much smaller host ranges for the others. The most abundant host species in ecotype II, III and IV were roe deer, rodents and birds, respectively (Table 5). Visual inspection of the haplotype genealogies within ecotype I indicates a mixture of A. phagocytophilum samples of all kind of vertebrate species and I. ricinus, indicating transmission of A. phagocytophilum between these host species via I. ricinus. The presence of ecotypes in European countries were plotted to test whether the clustering could be explained by differences in geographic distribution. All four ecotypes were spread over Europe, and no geographic clustering of the ecotypes was observed (Figure 2).
Geographic distributions of A. phagocytophilum ecotypes in Europe. Countries in which one or more isolates from an ecotype are found are filled with grey. A country in which an ecotype was not detected or which was not sampled is depicted in white. Data are based on isolates from Table 5. Number of isolates per country can be found in Additional file 3: Table S2.
Considerable sequence variation was found between and within ecotypes I, II and III. This prompted us to investigate whether this genetic marker could be used to detect changes in the population dynamics of A. phagocytophilum in the wild. The Ewens-Watterson test was performed separately on ecotypes I, II and III to infer the neutrality of the groEL marker [49–51]. It was not possible to apply the neutrality test for ecotype IV because only one haplotype was identified in this ecotype (Figure 1). The probability that two randomly chosen samples share the same haplotype (the F-values) agreed to the expectation of neutrality for ecotype III, but not ecotype I and II indicating that a particular haplotype was over-represented within each ecotype. Population expansion was tested using estimates of Fu’s Fs and Tajima’s D. Estimates of Fu’s Fs were significantly negative for ecotype I from the Netherlands and ecotype I from Europe (Table 7). These results demonstrate a large excess of rare genetic variants, over the expected genetic variants under the hypothesis of neutral selection and constant population size.
Discussion
This study addressed the circulation of A. phagocytophilum in ticks and vertebrates. Our investigations only detected the DNA of this microorganism and not their viability or infectivity. However, previous studies implicate I. ricinus ticks as vectors and all investigated animals as potential hosts for A. phagocytophilum[4]. Therefore, the inability of a DNA-based detection method to asseverate infectiousness in host species is expected to be a minor issue in this study. In any case, we revealed the widespread circulation of A. phagocytophilum in enzootic cycles in Belgium and the Netherlands. In some cases, the infection status of hosts was inferred from the infection rate of infected tick feeding. Vertebrates were considered positive when the infection rate of engorged ticks was significantly higher than that of questing I. ricinus (Table 4).
The absence of A. phagocytophilum in questing ticks in four out of 17 areas might be attributable to the relatively low number of ticks collected and tested (Table 1). Still, infection rates of A. phagocytophilum in questing ticks varied significantly between some geographic locations (Table 1), corroborating results from previous studies from other locations in Europe [4]. Knowledge on the ecological factors driving these differences is of relevance to public and animal health [52], but was not in the scope of this study. Significant differences between the infection rates of I. ricinus nymphs and adults were observed as well (Table 2). This may reflect that A. phagocytophilum ecotypes I and II cycle mainly between (infected) adults and nymphal I. ricinus,, which become infected when feeding on larger vertebrates.
Also, in another study a small but significant proportion of I. ricinus larvae were found A. phagocytophilum-positive [53]. These larvae may have become positive due to transovarial transmission or due to drop-off from A. phagocytophilum-positive hosts after partial feeding and continued to quest as larvae. Transovarial transmission and whether A. phagocytophilum-positive larvae can transmit the microorganism to vertebrate hosts needs to be investigated. Together, these findings indicate that all three tick stages should be taken into account when calculating the acarological risk of a given area [12].
In terms of the risk for public health, not only the product of the density of questing ticks and their infection rate defines high or low risk areas, but also the zoonotic potential of the microorganism should be taken into account [12]. The identification of four different A. phagocytophilum ecotypes (Figures 1 and 2) with significantly different host ranges and zoonotic potential supports this. A significant correlation between the genetic clustering of groEL sequences and different host ranges was found (Tables 5 and 6). Further genetic subclustering within ecotypes I, II and IV was also observed (Additional file 2: Figure S1). These subclusters could not be statistically linked to a further restriction in host ranges or to limitations in geographic distributions (not shown), probably due to lack of resolution in the groEL locus, and due to the limitations in the number and origin of the used samples, particularly of rodents and birds.
Clustering of A. phagocytophilum isolates can also be achieved using other genetic loci, such as the ankA gene, which is presumably involved in host-specific adaptation [21, 54]. Combining several genetic loci, such as groEL and AnkA, in future analyses could reveal more refined host ranges, especially within ecotype I. Recently, a multilocus sequence typing scheme for A. phagocytophilum was presented, which was shown to be informative concerning host species, geographic distribution, and zoonotic potential [54]. The advent of this standardized multilocus sequence typing scheme and a freely available molecular epidemiological database (http://pubmlst.org/aphagocytophilum/) will facilitate more elaborate analyses in the future.
Ecotype I had the broadest host range, but lacked birds and rodents, indicating that the latter two do not contribute directly to the transmission cycle. The generalist feeding behavior of I. ricinus nymphs and adults probably facilitates the continuous exchange of ecotype I between the different vertebrate species. All human isolates on the groEL-gene from Genbank and the literature [38–42] belong to ecotype I, demonstrating that members of this ecotype are zoonotic. Hence, ecotype I is the most plausible cause of infection regarding the one case of Human Granulocytic Anaplasmosis (HGA) in the Netherlands reported in 1999 [55]. Whether all or only a subset of the members of ecotype I are zoonotic remains to be examined [56].
In this study, ecotype II was found in roe deer, I. ricinus, and deer keds (Table 5). Therefore, ecotype II may circulate between roe deer via I. ricinus, or deer keds, or both. Whether deer keds may act as a host specific vector for ecotype II remains to be investigated [57, 58]. When the generalist tick I. ricinus would transmit ecotype II, then the observed host specificity might be attributed to A. phagocytophilum and the possibility that the vectors play a role in host specificity could be excluded. Only three isolates belonging to ecotype III were found in this study. One isolate was found in the spleen of a wood mouse. The kidney, liver and ear of this rodent were all A. phagocytophilum-positive, indicating a systemic infection of this wood mouse with A. phagocytophilum. Two isolates were found in two different tick species, I. trianguliceps and I. ricinus, feeding on one wood mouse. Ecotype III was not found in questing I. ricinus or in any other wildlife, except rodents (Table 5). Our finding supports the notion that ecotype III might be adapted to a life cycle involving exclusively some rodent species and a rodent specific vector, such as I. trianguliceps[18, 23, 59, 60]. Ecotype IV is most likely associated with one or more bird species, but not with other vertebrates. Ecotype IV was not found or in any other animal species. As ecotype IV was not found in questing I. ricinus either, it might be adapted to a life cycle involving exclusively birds and a bird-specific vector, such as I. frontalis. Although A. phagocytophilum was not detected in questing D. reticulatus or I. arboricola ticks, their role in the transmission of one or more A. phagocytophilum ecotypes cannot be excluded due to the relatively low numbers of ticks tested [61]. Although many ticks and animal samples have been included in this study, some animal species, particularly birds, rodents and carnivores, and some geographical locations (Figure 2) are underrepresented. Future studies should include broader and randomized sampling strategies.
Before the considerable sequence variation between and within ecotype I and II in the groEL gene (Figure 1) could be used to address their population dynamics, several statistical tests were performed to address the neutrality of this genetic marker. The Ewens-Watterson test detected significant departure from neutrality for ecotype I. This outcome indicated that a particular haplotype was identified in ecotype I more frequently than the neutral expectation, indicating that this haplotype is under positive selection. Fu’s Fs statistic detected genetic traces of demographic changes for ecotype I in the Netherlands and Belgium. Fu’s Fs is more sensitive than Tajima’s D to an excess of rare genetic variants in the samples [62], and this has proved to be true for our datasets (Table 7). The increase in ecotype I population sizes might have occurred through an increase in either the population of ixodid ticks, or in the vertebrate host species, or in both [9, 11].
Conclusions
In conclusion, we identify the groEL gene as a suitable marker to discriminate between A. phagocytophilum ecotypes. These ecotypes can be linked to distinct host ranges. Furthermore, all three ecotypes have enzootic cycles in the Netherlands and Belgium. In these countries, ecotype I is expanding. This is probably caused by the increase in abundance (and activity) of their vertebrate hosts and vectors. Based on the analyses of the groEL marker, we infer that: 1. Ecotype I has the highest zoonotic potential, and 2. the acarological risk of exposure to A. phagocytophilum ecotype I has been increasing in time. However, future studies concerning the evolution, population dynamics, and ecology of naturally occurring A. phagocytophilum will shed light on identifying risks for public health.
References
Rikihisa Y: Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev. 2011, 24: 469-489.
Ismail N, Bloch KC, McBride JW: Human ehrlichiosis and anaplasmosis. Clin Lab Med. 2010, 30: 261-292.
Chen SM, Dumler JS, Bakken JS, Walker DH: Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994, 32: 589-595.
Stuen S, Granquist EG, Silaghi C: Anaplasma phagocytophilum–a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013, 3: 31-
Edouard S, Koebel C, Goehringer F, Socolovschi C, Jaulhac B, Raoult D, Brouqui P: Emergence of human granulocytic anaplasmosis in France. Ticks Tick Borne Dis. 2012, 3: 403-405.
Heyman P, Cochez C, Hofhuis A, van der Giessen J, Sprong H, Porter SR, Losson B, Saegerman C, Donoso-Mantke O, Niedrig M, Papa A: A clear and present danger: tick-borne diseases in Europe. Expert Rev Anti Infect Ther. 2010, 8: 33-50.
Tijsse E, Koopmans M, Sprong H: Tick-borne pathogen – reversed and conventional discovery of disease. Frontiers Public Health. 2014, 2: 1-8.
Parola P, Davoust B, Raoult D: Tick- and flea-borne rickettsial emerging zoonoses. Vet Res. 2005, 36: 469-492.
Sprong H, Hofhuis A, Gassner F, Takken W, Jacobs F, van Vliet AJ, van Ballegooijen M, van der Giessen J, Takumi K: Circumstantial evidence for an increase in the total number and activity of Borrelia-infected Ixodes ricinus in the Netherlands. Parasit Vectors. 2012, 5: 294-
Hofhuis A, Herremans T, Notermans DW, Sprong H, Fonville M, van der Giessen JW, van Pelt W: A prospective study among patients presenting at the general practitioner with a tick bite or erythema migrans in The Netherlands. PLoS One. 2013, 8: e64361-
Coipan EC, Fonville M, Tijsse-Klasen E, van der Giessen JW, Takken W, Sprong H, Takumi K: Geodemographic analysis of Borrelia burgdorferi sensu lato using the 5S-23S rDNA spacer region. Infect Genet Evol. 2013, 17: 216-222.
Coipan EC, Jahfari S, Fonville M, Maassen CB, van der Giessen J, Takken W, Takumi K, Sprong H: Spatiotemporal dynamics of emerging pathogens in questing Ixodes ricinus. Front Cell Infect Microbiol. 2013, 3: 36-
Medlock JM, Hansford KM, Bormane A, Derdakova M, Estrada-Pena A, George JC, Golovljova I, Jaenson TG, Jensen JK, Jensen PM, Kazimirova M, Oteo JA, Papa A, Pfister K, Plantard O, Randolph SE, Rizzoli A, Santos-Silva MM, Sprong H, Vial L, Hendrickx G, Zeller H, Van Bortel W: Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasit Vectors. 2013, 6: 1-
Jin H, Wei F, Liu Q, Qian J: Epidemiology and control of human granulocytic anaplasmosis: a systematic review. Vector Borne Zoonotic Dis. 2012, 12: 269-274.
Rar V, Golovljova I: Anaplasma, Ehrlichia, and "Candidatus Neoehrlichia" bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011, 11: 1842-1861.
Cohan FM: Bacterial species and speciation. Syst Biol. 2001, 50: 513-524.
Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, Birtles RJ: High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol. 2007, 45: 1771-1776.
Bown KJ, Lambin X, Ogden NH, Begon M, Telford G, Woldehiwet Z, Birtles RJ: Delineating Anaplasma phagocytophilum ecotypes in coexisting, discrete enzootic cycles. Emerg Infect Dis. 2009, 15: 1948-1954.
de la Fuente J, Atkinson MW, Naranjo V, de Mera IG F, Mangold AJ, Keating KA, Kocan KM: Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet Microbiol. 2007, 119: 375-381.
Casey AN, Birtles RJ, Radford AD, Bown KJ, French NP, Woldehiwet Z, Ogden NH: Groupings of highly similar major surface protein (p44)-encoding paralogues: a potential index of genetic diversity amongst isolates of Anaplasma phagocytophilum. Microbiology. 2004, 150: 727-734.
Scharf W, Schauer S, Freyburger F, Petrovec M, Schaarschmidt-Kiener D, Liebisch G, Runge M, Ganter M, Kehl A, Dumler JS, Garcia-Perez AL, Jensen J, Fingerle V, Meli ML, Ensser A, Stuen S, von Loewenich FD: Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J Clin Microbiol. 2011, 49: 790-796.
Massung RF, Owens JH, Ross D, Reed KD, Petrovec M, Bjoersdorff A, Coughlin RT, Beltz GA, Murphy CI: Sequence analysis of the ank gene of granulocytic ehrlichiae. J Clin Microbiol. 2000, 38: 2917-2922.
Majazki J, Wuppenhorst N, Hartelt K, Birtles R, von Loewenich FD: Anaplasma phagocytophilum strains from voles and shrews exhibit specific ankA gene sequences. BMC Vet Res. 2013, 9: 235-
Sumner JW, Nicholson WL, Massung RF: PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J Clin Microbiol. 1997, 35: 2087-2092.
Carpi G, Bertolotti L, Pecchioli E, Cagnacci F, Rizzoli A: Anaplasma phagocytophilum groEL gene heterogeneity in Ixodes ricinus larvae feeding on roe deer in Northeastern Italy. Vector Borne Zoonotic Dis. 2009, 9: 179-184.
Herbeck JT, Funk DJ, Degnan PH, Wernegreen JJ: A conservative test of genetic drift in the endosymbiotic bacterium Buchnera: slightly deleterious mutations in the chaperonin groEL. Genetics. 2003, 165: 1651-1660.
Rymaszewska A: Divergence within the marker region of the groESL operon in Anaplasma phagocytophilum. Eur J Clin Microbiol Infect Dis. 2008, 27: 1025-1036.
Silaghi C, Hamel D, Thiel C, Pfister K, Passos LM, Rehbein S: Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis. 2011, 11: 355-362.
Jahfari S, Fonville M, Hengeveld P, Reusken C, Scholte EJ, Takken W, Heyman P, Medlock J, Heylen D, Kleve J, Sprong H: Prevalence of Neoehrlichia mikurensis in ticks and rodents from North-west Europe. Parasit Vectors. 2012, 5: 74-
Heylen D, Tijsse E, Fonville M, Matthysen E, Sprong H: Transmission dynamics of Borrelia burgdorferi s.l. in a bird tick community. Environ Microbiol. 2013, 15: 663-673.
Pacilly F, Benning A, Jacobs F, Leidekker J, Sprong H, van Wieren SE, Takken W: Blood feeding on large grazers affects the transmission of Borrelia burgdorferi sensu lato by Ixodes ricinus. Ticks and Tick-borne Dis. 2014, Accepted
Tijsse-Klasen E, Fonville M, Reimerink JH, van der Spitzen Sluijs A, Sprong H: Role of sand lizards in the ecology of Lyme and other tick-borne diseases in the Netherlands. Parasit Vectors. 2010, 3: 42-
Franssen F, Nijsse R, Mulder J, Cremers H, Dam C, Takumi K, van der Giessen J: Increase in number of helminth species from Dutch red fox over a 35-year period. Parasit Vectors. 2014, 7: 166-
Butler CM, Nijhof AM, Jongejan F, van der Kolk JH: Anaplasma phagocytophilum infection in horses in the Netherlands. Vet Rec. 2008, 162: 216-217.
Courtney JW, Kostelnik LM, Zeidner NS, Massung RF: Multiplex real-time PCR for detection of anaplasma phagocytophilum and Borrelia burgdorferi. J Clin Microbiol. 2004, 42: 3164-3168.
Alberti A, Zobba R, Chessa B, Addis MF, Sparagano O, Pinna Parpaglia ML, Cubeddu T, Pintori G, Pittau M: Equine and canine Anaplasma phagocytophilum strains isolated on the island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl Environ Microbiol. 2005, 71: 6418-6422.
Foldvari G, Jahfari S, Rigo K, Jablonszky M, Szekeres S, Majoros G, Toth M, Molnar V, Coipan EC, Sprong H: Candidatus Neoehrlichia mikurensis and Anaplasma phagocytophilum in Urban Hedgehogs. Emerg Infect Dis. 2014, 20: 496-498.
Lotric-Furlan S, Rojko T, Petrovec M, Avsic-Zupanc T, Strle F: Epidemiological, clinical and laboratory characteristics of patients with human granulocytic anaplasmosis in Slovenia. Wien Klin Wochenschr. 2006, 118: 708-713.
Arnez M, Petrovec M, Lotric-Furlan S, Zupanc TA, Strle F: First European pediatric case of human granulocytic ehrlichiosis. J Clin Microbiol. 2001, 39: 4591-4592.
Haschke-Becher E, Bernauer R, Walleczek AM, Apfalter P, Afazel-Saeedi S, Kraus J, Ladurner G, Strasser P: First detection of the Anaplasma phagocytophilum groEL-A genotype in man. J Infect. 2010, 60: 300-305.
Smrdel KS, Petrovec M, Furlan SL, Zupan TA: The sequences of groESL operon of Anaplasma phagocytophilum among human patients in Slovenia. FEMS Immunol Med Microbiol. 2012, 64: 123-125.
Petrovec M, Sumner JW, Nicholson WL, Childs JE, Strle F, Barlic J, Lotric-Furlan S, Avsic Zupanc T: Identity of ehrlichial DNA sequences derived from Ixodes ricinus ticks with those obtained from patients with human granulocytic ehrlichiosis in Slovenia. J Clin Microbiol. 1999, 37: 209-210.
Cochez C, Ducoffre G, Vandenvelde C, Luyasu V, Heyman P: Human anaplasmosis in Belgium: a 10-year seroepidemiological study. Ticks Tick Borne Dis. 2011, 3: 156-159.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690.
Lanfear R, Calcott B, Ho SY, Guindon S: Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012, 29: 1695-1701.
Katoh K, Misawa K, Kuma K, Miyata T: MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30: 3059-3066.
Excoffier L, Lischer HE: Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010, 10: 564-567.
Salzburger W, Ewing GB, Von Haeseler A: The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol Ecol. 2011, 20: 1952-1963.
Ewens WJ: The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972, 3: 87-112.
Slatkin M: An exact test for neutrality based on the Ewens sampling distribution. Genet Res. 1994, 64: 71-74.
Watterson GA: Heterosis or neutrality?. Genetics. 1977, 85: 789-814.
Braks M, van der Giessen J, Kretzschmar M, van Pelt W, Scholte EJ, Reusken C, Zeller H, van Bortel W, Sprong H: Towards an integrated approach in surveillance of vector-borne diseases in Europe. Parasit Vectors. 2011, 4: 192-
Krucken J, Schreiber C, Maaz D, Kohn M, Demeler J, Beck S, Schein E, Olias P, Richter D, Matuschka FR, Pachnicke S, Krieger K, Kohn B, von Samson-Himmelstjerna G: A novel high-resolution melt PCR assay discriminates Anaplasma phagocytophilum and "Candidatus Neoehrlichia mikurensis". J Clin Microbiol. 2013, 51: 1958-1961.
Huhn C, Winter C, Wolfsperger T, Wuppenhorst N, Strasek Smrdel K, Skuballa J, Pfaffle M, Petney T, Silaghi C, Dyachenko V, Pantchev N, Straubinger RK, Schaarschmidt-Kiener D, Ganter M, Aardema ML, von Loewenich FD: Analysis of the population structure of Anaplasma phagocytophilum using multilocus sequence typing. PLoS One. 2014, 9: e93725-
van Dobbenburgh A, van Dam AP, Fikrig E: Human granulocytic ehrlichiosis in western Europe. N Engl J Med. 1999, 340: 1214-1216.
Barbet AF, Al-Khedery B, Stuen S, Granquist EG, Felsheim RF, Munderloh UG: An Emerging Tick-Borne Disease of Humans Is Caused by a Subset of Strains with Conserved Genome Structure. Pathogens. 2013, 2: 544-555.
Zaugg JL, Coan ME: Test of the sheep ked Melophagus ovinus (L) as a vector of Anaplasma ovis Lestoquard. Am J Vet Res. 1986, 47: 1060-1062.
Víchová B, Majláthová V, Nováková M, Majláth I, Curlík J, Bona M, Komjáti-Nagyová M, Peťko B: PCR detection of re-emerging tick-borne pathogen, Anaplasma phagocytophilum, in deer ked (Lipoptena cervi) a blood-sucking ectoparasite of cervids. Biologia. 2011, 66: 1082-1086.
Bown KJ, Begon M, Bennett M, Birtles RJ, Burthe S, Lambin X, Telfer S, Woldehiwet Z, Ogden NH: Sympatric Ixodes trianguliceps and Ixodes ricinus ticks feeding on field voles (Microtus agrestis): potential for increased risk of Anaplasma phagocytophilum in the United Kingdom?. Vector Borne Zoonotic Dis. 2006, 6: 404-410.
Rar VA, Epikhina TI, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, Ivanov LI: Genetic variability of Anaplasma phagocytophilum in Ixodes persulcatus ticks and small mammals in the Asian part of Russia. Vector Borne Zoonotic Dis. 2011, 11: 1013-1021.
Richter D, Kohn C, Matuschka FR: Absence of Borrelia spp., Candidatus Neoehrlichia mikurensis, and Anaplasma phagocytophilum in questing adult Dermacentor reticulatus ticks. Parasitol Res. 2013, 112: 107-111.
Fu YX: Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997, 147: 915-925.
Acknowledgements
The authors are very grateful to the volunteers of animal-shelters, particularly to Jenny Kleve for her dedicated work on hedgehogs. We thank the Centre Monitoring Vectors for collecting tick, Margriet Montizaan for collecting deer keds and Chantal Reusken for sharing rodent materials. This study was financially supported by the Dutch Food and Consumer Product Safety Authority (NVWA), Wageningen University and Research Centre (WUR) and by the Ministry of Health, Welfare and Sport (VWS). G.F. was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. The work of CC, PHe, DH and HS was done under the frame of EurNegVec Cost Action TD1303.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SJ, WTa and HS designed parts of the study. DH, PHe, CM, CB, JR, GD, GF, SSz, GD, WT, SW and JG collected wildlife samples. SJ, MF, AL and PH performed lab tests and laboratory analyses. SJ, CC, KT and HS analyzed data and performed statistical and population genetic analysis. WTa and HS supervised different parts of study. SJ and HS wrote the final manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13071_2014_1562_MOESM2_ESM.pdf
Additional file 2: Figure S1: Phylogenetic relationship of A. phagocytophilum. Phylogenetic analyses of groEL sequences from all A. phagocytophilum samples (Table 5) were performed as described in the Methods section. Roman numerals label the four ecotypes. (PDF 41 KB)
13071_2014_1562_MOESM3_ESM.doc
Additional file 3: Table S2: Geographic distributions of A. phagocytophilum ecotypes in Europe. Number of isolates per country. Data are based on isolates from Table 3. (DOC 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Jahfari, S., Coipan, E.C., Fonville, M. et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasites Vectors 7, 365 (2014). https://doi.org/10.1186/1756-3305-7-365
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-7-365