Abstract
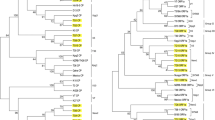
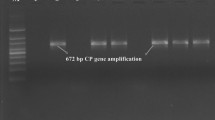
The present study for the first time describes biological and molecular characterization of Citrus tristeza virus (CTV) occurring in the Terai area of Uttarakhand State in Northern Himalaya region of India. Direct antigen coated-ELISA and reverse transcriptase-polymerase chain reaction (RT-PCR) detected the CTV infection in Acid lime cv. Pant lemon (Citrus aurantifolia) orchards of Pantnagar with an estimated disease incidence of 16.6–20.5 %. To know the biological and genetic properties, an isolate, CTV Pant 4 was characterized. Isolate Pant 4 could be graft transmitted to Kinnow, Nagpur and Darjeeling mandarins, Mosambi sweet orange, Kagzi lime, Sweet lime, Sour orange but not to Rough lemon. The sequence analyses of the 5′ORF1a (3038 nucleotides) of LPro domain and 3′end (2058 nt) covering ORF7–ORF10 regions of the CTV genome revealed that Pant 4 was closely related to the previously reported Indian CTV isolate, Kpg3 from Northeastern Himalaya region with 97 and 98 % sequence identity, respectively. Whereas, it differed from the previously reported CTV isolate B165 from Southern India with 79 and 92 % identity, respectively for 5′ORF1a and 3′ end regions. Recombination and SplitsTree decomposition analyses indicated that CTV isolate Pant 4 was a recombinant isolate originating from Kpg3 as a major and B165 as a minor donor.


Similar content being viewed by others
References
Ahlawat YS. Viruses, greening bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agric Sci. 1997;67:51–7.
Ahlawat YS, Pant RP. Major virus and virus-like diseases of citrus in India: their diagnosis and management. Ann Rev Plant Pathol. 2003;2:447–74.
Bar-Joseph M, Dawson WO. Citrus tristeza virus. Encyclopedia Virol. 2008;1:520–5.
Biswas KK. Molecular diagnosis of Citrus tristeza virus in mandarin (Citrus reticulata) orchards of hills of West Bengal. Indian J Virol. 2008;19:26–31.
Biswas KK. Molecular characterization of Citrus tristeza virus isolates from the Northeastern Himalayan region of India. Arch Virol. 2010;155:959–63.
Biswas KK, Tarafdar A, Sharma SK. Complete genome of mandarin decline Citrus tristeza virus of Northeastern Himalayan hill region of India: comparative analyses determine recombinant. Arch Virol. 2012;157:579–83.
Biswas KK, Tarafdar A, Diwedi S, Lee RF. Distribution, genetic diversity and recombination analysisof Citrus tristeza virus of India. Virus Genes. 2012;45:139–48.
Cheng X, Wu X, Wang H, Sun Y, Qian Y, Luo L. High codon adaptation in Citrus tristeza virus to its citrus host. Virol J. 2012;9:113.
Ghosh DK, Aghave B, Roy A, Ahlawat YS. Molecular cloning, sequencing and phylogenetic analysis of coat protein gene of a biologically distinct CTV isolate occurring in central India. J Plant Biochem Biotechnol. 2009;18:105–8.
Harper SJ, Dawson TE, Pearson MN. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol. 2010;155:471–80.
Hilf ME, Mavrodieva VA, Garnsey SM. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology. 2005;95:909–17.
Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, Niblett CL, Cline K, Gumpf DJ, Lee RF. Complete sequence of the Citrus tristeza virus RNA genome. Virology. 1995;208:511–20.
Korkmaz S, Cevik B, Onder S, Koc NK. Biological, serological and molecular characterization of Citrus tristeza virus isolates from different citrus cultivation regions of Turkey. Turk J Agric. 2008;32:369–79.
Lee RF, Bar-Joseph M. Tristeza. In: Compendium of citrus diseases, 2nd ed. St. Paul, MN, USA, APS Press; 2000. pp 61–3.
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P. Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol. 2009;90:1527–38.
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS. Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes. 2010;40:111–8.
Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol. 2008;9(2):251–68.
Rocha-Pena MA, Lee RF, Lastra R, Niblett CL, Ochoa-Corona FM, Garnsey SM, Yokomi RK. Citrus tristeza virus and its aphid vector Toxoptera citricida: threats to citrus production in the Caribbean and Central and North America. Plant Dis. 1995;79:437–45.
Roy A, Brlansky RH. Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res. 2010;151:118–30.
Roy A, Manjunath KL, Brlansky RH. Assessment of sequence diversity in the 5-terminal region of Citrus tristeza virus from India. Virus Res. 2005;113:132–42.
Satyanayanana T, Gowda S, Boyko VP, Albiach-Marti MR, Mawassi M, Navas-Castillo J, Karasev AV, Dolja V, Hilf ME, Lewandowsky DJ, Moreno P, Bar-Joseph M, Garnsey SM, Dawson WO. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci U S A. 1999;96:7433–8.
Satyanayanana T, Gowda S, Mawassi M, Albiach-Marti MR, Ayllon MA, Robertson C, Garnsey SM, Dawson WO. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology. 2000;278:253–65.
Sharma SK, Tarafdar A, Khatun D, Kumari S, Biswas KK. Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus isolates in Delhi region of India. J Plant Biochem Biotechnol. 2012;21:38–43.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9.
Tarafdar A, Ghosh PD, Biswas KK. In planta distribution, accumulation, movement and persistence of Citrus tristeza virus in citrus host. Indian Phytopathol. 2012;65:184–8.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82.
Acknowledgments
The authors are grateful to Head, Division of Plant Pathology; Director, IARI, New Delhi for providing financial and laboratory support; Prof. Anupam Varma for his guidance in this work. This study has been supported by Research Project (Code No. 24-33), Department of Biotechnology, Govt. of India. The first author is grateful to ICAR for financial assistance as JRF and thankful to Dr. A. K. Singh and Dr. (Mrs.) P. Sharma, IARI, for valuable suggestions. We are thankful to N. Trivedi, GBPAU&T, Pantnagar for his assistance during survey and sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, J.K., Tarafdar, A., Sharma, S.K. et al. Evidence of Recombinant Citrus tristeza virus Isolate Occurring in Acid Lime cv. Pant Lemon Orchard in Uttarakhand Terai Region of Northern Himalaya in India. Indian J. Virol. 24, 35–41 (2013). https://doi.org/10.1007/s13337-012-0118-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-012-0118-8