Abstract
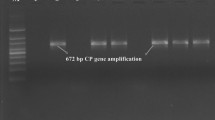
Infection of Citrus tristeza virus (CTV) in different citrus orchards of New Delhi was detected by direct antigen coated-ELISA and RT-PCR. Sweet orange (Citrus sinensis) orchards were found to be susceptible to CTV with estimated disease incidence up to 39%. Kagzi kalan (C. lemon), Pumello (C. paradisi) and Kinnow mandarin (C. reticulata) orchards did not show CTV infection. Three CTV isolates, D1, D7 and D15 randomly selected from infected sweet orange orchards were considered for biological and molecular characterization. In the host range study, all the Delhi isolates infected Darjeeling mandarin (C. reticulata), Kagzi lime (C. aurantifolia), sour orange (C. aurantium) and sweet orange but not Kinnow mandarin. A fragment of 5′ORF1a and complete coat protein (CP) gene of these three isolates were cloned, sequenced and compared with other Indian and international CTV isolates. Delhi isolates shared 85–92% sequence identity for 5′ORF1a fragment and 89–91% for CP gene among them. Phylogenetic analysis segregated three Delhi isolates into three genogroups for each of 5′ORF1a fragment and CP gene, however phylogenetic relationships for both the genomic regions was incongruent. Recombination detecting program RDP3 detected CTV isolate D7 as recombinant, indicating genetic variability in CTV isolates might be the outcome of recombination events between divergent CTV sequences. An attempt was made in present study to characterize CTV isolates biologically and at genetic level, and to determine genetic diversity at farm level and study the recombination of CTV isolates in Delhi region.


Similar content being viewed by others
Abbreviations
- cDNA:
-
Complimentary deoxyribo nucleic acid
- CP:
-
Coat protein
- CTV:
-
Citrus tristeza virus
- ELISA:
-
Enzyme-linked immunosorbent assay
- kb:
-
Kilo base
- NCBI:
-
National centre for biotechnology information
- nt:
-
Nucleotide
- ORF:
-
Open reading frame
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Ahlawat YS (1997) Viruses, greening bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agr Sci 67:51–57
Bar-Joseph M, Dawson WO (2008) Citrus tristeza virus. Encyclopedia of Virology pp 520–525
Biswas KK (2008) Molecular diagnosis of Citrus tristeza virus in mandarin (Citrus reticulata) orchards of hills of West Bengal. Indian J Virol 19:26–31
Biswas KK (2010) Molecular characterization of Citrus tristeza virus isolates from the Northeastern Himalayan region of India. Arch Virol 155:959–963
Chakraborty NK, Ahlawat YS, Varma A, Chandra KJ, Ramapandu S, Kapur SP (1992) Serological reactivity in Citrus tristeza virus strains in India. In: Proc 12th Conf IOCV, Riverside, CA, USA, pp 108–112
Hilf ME, Mavrodieva VA, Garnsey SM (2005) Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopath 95:909–917
Lee RF, Bar-Joseph M (2000) Tristeza. In: Timmer LW, Garnsey SM, Graham JH (eds) Compendium of citrus diseases, 2nd edn, APS. APS, St Paul, pp 61–63
Lee RF, Garnsey SM (1996) Citrus virus and virus like pathogens: a continuous evolution of progress and problems. In: Proc. 13th Conf. Intern. Organ Citrus Virol China, IOCV, Riverside, California, pp 1–7
Martin DP (2009) Recombination detection and analysis using RDP3. Meth Mol Biol 537:185–205
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P (2009) Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol 90:1527–1538
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS (2010) Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Gene 40:111–118
Roy A, Brlansky RH (2010) Genome of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Roy A, Manjunath KL, Brlansky RH (2005) Assessment of sequence diversity in the 5-terminal region of Citrus tristeza virus from India. Virus Res 113:132–142
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Sambrook J, Fritch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 24:4876–4882
Acknowledgements
First author is grateful to ICAR for financial assistantship as JRF. This work was financially supported through externally funded Research Project of Department of Biotechnology, Govt. of India (Project code 24:33). Authors are grateful to Director, IARI; Dr. R.K. Jain, Head, Division of Plant Pathology; Dr. V.G. Malathi, In-charge, ACPV, IARI for providing financial and laboratory facilities. First author is also grateful to Dr. T.R. Sharma and Dr. S.C. Dubey, IARI for their valuable suggestions during this research work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 3
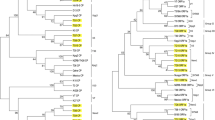
UPGMA phylogenetic tree analysis in RDP3; For non-recombinant (385–234 nt) (a) and recombinant part (235–384 nt) (b) in 5 ORF1a; non-recombinant (154–449 nt) (c) and recombinant part (450–153 nt) (d) in CP sequences. Parental sequences are in bold and recombinant in underline. (PDF 27 kb)
Rights and permissions
About this article
Cite this article
Sharma, S.K., Tarafdar, A., Khatun, D. et al. Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus in Delhi region of India. J. Plant Biochem. Biotechnol. 21, 38–43 (2012). https://doi.org/10.1007/s13562-011-0071-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-011-0071-4