Abstract
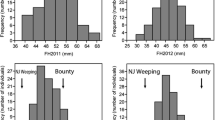
Two intraspecific peach breeding populations have been used to conduct a quantitative trait locus (QTL) analysis of fruit quality traits: an F1 from the cross Bolero (B) x OroA (O) and an F2 from the cross Contender (C) x Ambra (A). A total of 344 Prunus simple sequence repeats (SSRs) were analyzed in B, O, C, A parents and CxA F1 hybrid. Eight SSR were mapped for the first time in peach. A multiplex-ready polymerase chain reaction (PCR) protocol has allowed considerable time and cost saving during genotyping steps. Two maps (B map and O map) were produced for BxO population following the pseudo-test cross strategy and one for CxA. No marker could be mapped on G6 for the B map, on G4 and G8 for the O map and on G5 for the CxA map. Both populations were phenotyped over 2 years for maturity date (MD), fruit weight, external fruit skin overcolor, juice total soluble solids (SSC, Brix degree), juice titrable acidity and juice pH. Data for blooming time and flower type were scored only for BxO in 2007. All traits had a normal distribution, except for MD which was bimodal in BxO and trimodal in CxA, where it was scored as a co-dominant trait. Up to two QTLs per trait were detected in each population, and most of them were located in the same region forming clusters of QTLs, especially on G4. This is likely due to a major pleiotropic effect of MD masking the identification of other QTLs for different traits.


Similar content being viewed by others
References
Abbott A, Rajapaske S, Sosinski B, Lu ZX, Sossy-Alaoui K, Gannavarapu M, Reighard G, Ballard RE, Baird WV, Scorza R, Callahan A (1998) Construction of saturated linkage maps of peach crosses segregating for characters controlling fruit quality, tree architecture and pest resistance. Acta Hort 465:41–49
Abbott AG, Arús P, Scorza R (2008) Genetic engineering and genomics. In: Desmond RL, Bassi D (eds) The peach: botany, production and uses. CABI, UK, pp 85–105
Aranzana MJ, Pineda A, Cosson P, Ascasibar J, Dirlewanger E, Ascasibar J, Cipriani G, Ryder CD, Testolin R, Abbott A, King GJ, Iezzoni AF, Arùs P (2003) A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor Appl Genet 106:819–825
Arús P, Yamamoto T, Dirlewanger E, Abbott AG (2005) Synteny in the Rosaceae. In: Janick J (ed) Plant Breeding Reviews. Wiley, London, pp 175–211
Arús P, Yamamoto T, Dirlewanger E, Abbott AG (2006) Synteny in the Rosaceae. Plant Breed Rev 27: 175–211
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Bassi D, Gambardella M, Negri P (1988) Date of ripening and two morphological fruit traits in peach progenies. Acta Hort 254:59–66
Beckman TG, Sherman WB (2003) Probable qualitative inheritance of full red skin color in peach. HortScience 38:1184–1185
Beckman TG, Rodriguez AJ, Sherman WB, Werner DJ (2005) Evidence for qualitative suppression of red skin color in peach. HortScience 40:523–524
Blenda AV, Verde I, Georgi LL, Reighard GL, Forrest SD, Muñoz-Torres M, Baird WV, Abbott A (2007) Construction of a genetic linkage map and identification of molecular markers in peach rootstocks for response to peach tree short life syndrome. Tree Genet Genomes 3:341–350
Boudehri K, Bendahmane A, Cardinet G, Troadec C, Moing A, Dirlewanger E (2009) Phenotypic and fine genetic characterization of the D locus controlling fruit acidity in peach. BMC Plant Biol 9:59
Cantini C, Iezzoni AF, Lamboy WF, Boritzki M, Struss D (2001) DNA fingerprinting of tetraploid cherry germplasm using simple sequence repeats. J Amer Soc Hort Sci 126:205–209
Cipriani G, Lot G, Huang WG, Matarazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: Isolation, characterisation and cross species amplification in Prunus. Theor Appl Genet 99:65–72
Clarke JB, Tobutt KR (2003) Development and characterization of polymorphic microsatellites from Prunus avium ‘Napoleon’. Mol Ecol Notes 3:578–580
Clarke J, Sargent D, Bošković R, Belaj A, Tobutt K (2009) A cherry map from the interspecific cross Prunus avium ‘Napoleon’ × P. nipponica based on microsatellite, genespecific and isoenzyme markers. Tree Genet Genomes 5:41–51
Darvasi A, Weinreb A, Minke V, Weller JI, Soller M (1993) Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic-map. Genetics 134:943–951
Dirlewanger E, Pascal T, Zuger C, Kervella J (1996) Analysis of molecular markers associated with powdery mildew resistance genes in peach [Prunus persica (L.) Batsch] x Prunus davidiana hybrids. Theor Appl Genet 93:909–919
Dirlewanger E, Pronier V, Parvery C, Rothan C, Guy A, Monet R (1998) Genetic linkage map of peach [Prunus persica (L.) Batsch] using morphological and molecular markers. Theor Appl Genet 97:888–895
Dirlewanger E, Moing A, Rothan C, Svanella L, Pronier V, Guye A, Plomion C, Monet C (1999) Mapping QTLs controlling fruit quality in peach. Theor Appl Genet 98:18–31
Dirlewanger E, Cosson P, Tavaud M, Aranzana J, Poizat C, Zanetto A, Arus P, Laigret F (2002) Development of microsatellite markers in peach [ Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry ( Prunus avium L.). Theor Appl Genet 105:127–138
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 29:9891–9896
Dirlewanger E, Cosson P, Boudehri K, Renaud C, Capdeville G, Tauzin Y, Laigret F, Moing A (2006) Development of a second-generation genetic linkage map for peach [Prunus persica (L.) Batsch] and characterization of morphological traits affecting flower and fruit. Tree Genetics Genomes 3:1–13
Downey SL, Iezzoni AF (2000) Polymorphic DNA markers in black cherry (Prunus serotina) are identified using sequences from sweet cherry, peach, and sour cherry. J Amer Soc Hort Sci 125:76–80
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transription factor, MdMYB10. Plant J 49:414–427
Etienne C, Rothan C, Moing A, Plomion C, Bodnes C, Svanella-Dumas L, Cosson P, Pronier V, Monet R, Dirlewanger E (2002) Candidate gene and QTLs for sugar and organic acid content in peach. Theor Appl Genet 105:145–159
Gasic K, Han Y, Kertbundit S, Shulaev V, Iezzoni A, Stover E, Bell R, Wisniewski M, Korban S (2009) Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Mol Breed 23:397–411
Hagen LS, Chaib J, Fady B, Decroocq V, Bouchet JP, Lambert P, Audergon JM (2004) Genomic and cDNA microsatellites from apricot (Prunus armeniaca L.). Mol Ecol Notes 4:742–745
Hayden MJ, Nguyen TM, Waterman A, Chalmers KJ (2008) Multiplex-ready PCR: a new method for multiplexed SSR and SNP genotyping. BMC Genomics 9:80
Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, Arús P (2005) Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics 171:1305–1309
Illa E, Lambert P, Quilot B, Audergon JM, Dirlewanger E, Howad W, Dondini L, Tartarini S, Lain O, Testolin R, Bassi D, Arús P (2009) Linkage map saturation, construction, and comparison in four populations of Prunus. J Hortic Sci Biotech 84:168–175
Jung S, Jiwan D, Cho I, Lee T, Abbott A, Sosinski B, Main D (2009) Synteny of Prunus and other model plant species. BMC Genomics 10:76
Kearsey MJ, Farquhar AGL (1998) QTL analysis in plants; where are we now? Heredity 80:137–142
Kenis K, Keulemans J, Davey MW (2008) Identification and stability of QTLs for fruit quality traits in apple. Tree Genetics Genome 4:647–661
Kim SH, Lee JR, Hong ST, Yoo YK, An G, Kim SR (2003) Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 165:403–413
Lopes MS, Sefc KM, Laimer M, Machado AD (2002) Identification of microsatellite loci in apricot. Mol Ecol Notes 2(1):24–26
Messina R, Lain O, Marrazzo MT, Cipriani G, Testolin R (2004) New set of microsatellite loci isolated in apricot. Mol Ecol Notes 4:432-434
Mnejja M, Garcia-Mas J, Howad W, Arus P (2005) Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol Ecol Notes 5:531–535
Monet R (1979) Genetic transmission of the “non-acid” character. Incidence on selection for quality. In: InRA (ed) Eucarpia symposium on tree fruit breeding, angers., pp 273–276
Monforte AJ, Asins MJ, Carbonell EA (1999) Salt tolerance in Lycopersicon spp. VII. Pleiotropic action of genes controlling earliness on fruit yield. Theor Appl Genet 98:593–601
Morgutti S, Negrini N, Nocito FF, Ghiani A, Bassi D, Cocucci M (2006) Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phytol 171:315–328
Ogundiwin EA, Peace CP, Nicolet CM, Rashbrook VK, Gradziel TM, Bliss FA, Parfitt D, Crisosto CH (2008) Leucoanthocyanidin dioxygenase gene (PpLDOX): a potential functional marker for cold storage browning in peach. Tree Genet Genomes 4:543–554. doi:10.1007/s11295-007-0130-0
Ogundiwin EA, Peace CP, Gradziel TM, Parfitt DE, Bliss FA, Crisosto CH (2009) A fruit quality gene map of Prunus. BMC Genomics 10:587
Peace CP, Crisosto CH, Gradziel TM (2005) Endopolygalacturonase: a candidate gene for freestone and melting flesh in peach. Mol Breed 16:21–31
Quarta R, Dettori MT, Sartori A, Verde I (2000) Genetic linkage map and QTL analysis in peach. Acta Hort 521:233–241
Quilot B, Wu BH, Kervella J, Genard M, Foulogne M, Moreau K (2004) QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P. davidiana. Theor Appl Genet 109:884–897
R Development Core Team (2008) R: a language and environment for statistical computing. Version 2.7.1. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org. Accessed January 2009
Rodriguez GA, Sherman WB (1990) ‘OroA’ peach germplasm. Hortscience 25:128
Sanchez-Perez R, Howad W, Dicenta F, Arus P, Martinez-Gomez P (2007) Mapping major genes and quantitative trait loci controlling agronomic traits in almond. Plant Breed 126:310–318
Sargent DJ, Fernàndez-Fernàndez F, Rys A, Knight VH, Simpson DW, Tobutt KR (2007) Mapping of A1 conferring resistance to the aphid Amphorophora idaei and dw (dwarfing habit) in red raspberry (Rubus idaeus L.) using AFLP and microsatellite markers. BMC Plant Biol 7:15
Scorza R, Mehlenbacher SA, Lightner GW (1985) Inbreeding and coancestry of freestone peach cultivars of the eastern United States and implications for peach germplasm improvement. J Am Soc Hortic Sci 110:547–552
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, Pancaldi M, Sansavini S (2000) Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43(3):512–520
Testolin R, Messina R, Lain O, Marrazzo MT, Huang WG, Cipriani G (2004) Microsatellites isolated in almond from an AC-repeat enriched library. Mol Ecol Notes 4:459–461
Van Ooijen JW (2006) JoinMap ® 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Van Ooijen JW (2009) MapQTL ® 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, Netherlands
Vaughan SP, Russell K (2004) Characterization of novel microsatellites and development of multiplex PCR for large-scale population studies in wild cherry, Prunus avium. Mol Ecol Notes 4:429–431
Vecchietti A, Lazzari B, Ortugno C, Bianchi F, Caprera A, Malinverni R, Mignani I, Pozzi C (2009) Comparative analysis of expressed sequence tags from tissues in ripening stages of peach (Prunus persica L.). Tree Genet Genomes 5:377–391
Verde I, Quarta R, Cedrola C, Dettori MT (2002) QTL analysis of agronomic traits in a BC1 peach population. Acta Hort 592:291–295
Verde I, Lauria M, Dettori MT, Vendramin E, Balconi C, Micali S, Wang Y, Marrazzo MT, Cipriani G, Hartings H, Testolin R, Abbott AG, Motto M, Quarta R (2005) Microsatellite and AFLP markers in the Prunus persica [L. (Batsch)]xP. ferganensis BC(1)linkage map: saturation and coverage improvement. Theor Appl Genet 11:1013–1021
Vilanova S, Sargent DJ, Arús P, Monfort A (2008) Synteny conservation between two distantly-related Rosaceae genomes: Prunus (the stone fruits) and Fragaria (the strawberry). BMC Plant Biol 8:67
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Xu Y, Ma RC, Xie H, Liu JT, Cao MQ (2004) Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 47:1091–1104
Xu Y, Zhang L, Xie H, Zhang YQ, Olivieira M, Ma RC (2008) Expression analysis and genetic mapping of three SEPALLATA -like genes from peach (Prunus persica (L.) Batsch). Tree Genetics & Genomes 4:693–703
Yamamoto T, Shimada T, Imai T, Yaegaki H, Haji T, Matsuta N, Yamaguchi M, Hayashi T (2001) Characterization of morphological traits based on a genetic linkage map in peach. Breed Sci 51:271–278
Yamamoto T, Yamaguchi M, Hayashi T (2005) An integrated genetic linkage map of peach by SSR, STS, AFLP and RAPD. J Jpn Soc Hort Sci 74:204–213
Acknowledgments
We wish to thank C. Buscaroli, S. Foschi, E. Olivi, and N. Mamini for their valuable contribution in field tree management, fruit harvesting, data collection, and partial processing and Prof. Salamini and Dr. John Williams (PTP, Lodi) for the critical reading of the manuscript. We also would like to acknowledge an anonymous referee for valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Abbott
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
(DOC 9545 kb)
Rights and permissions
About this article
Cite this article
Eduardo, I., Pacheco, I., Chietera, G. et al. QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genetics & Genomes 7, 323–335 (2011). https://doi.org/10.1007/s11295-010-0334-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0334-6