Abstract
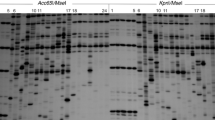
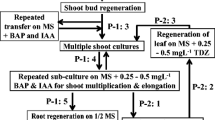
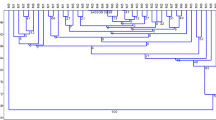
A simple tissue culture protocol was developed for efficient plant regeneration from young inflorescence-derived calli in wild barley, Hordeum brevisubulatum (Trin.) Link, an important pasturage grass. Genetic and epigenetic instabilities in the regenerated plants (regenerants) were assessed by three molecular markers AFLP, S-SAP and MSAP. Two pools of calli derived from young inflorescences of a single donor plant and 44 randomly chosen regenerants were subjected to AFLP analysis. Results showed that 74 out of 793 scored bands were polymorphic among the studied samples, giving rise to a genetic variation frequency of 9.3%. The number of variant bands as compared to the donor plant varied greatly among the regenerants, with a small number of regenerants accumulated a large number of variant bands (maximum 55), while the majority of regenerants showed only 2–3 variant bands. A subset of regenerants together with the two pools of calli were selected for S-SAP and MSAP analysis to detect possible retrotranspositional activity of a prominent retroelement family, BARE-1, in the genomes of Hordem species, and possible alterations in cytosine methylation. S-SAP analysis showed that of the 768 scored bands, 151 were polymorphic among the analyzed samples, giving rise to a genetic variation frequency of 19.7%, albeit no evidence for retrotranspositional event was obtained based on locus-specific PCR amplifications. MSAP analysis revealed that tissue culture has caused cytosine methylation alterations in both level and pattern compared with the donor plant. Sequencing of selected variant bands indicated that both protein-coding genes and transposon/retrotransposons were underlying the genetic and epigenetic variations. Correlation analysis of the genetic and epigenetic instabilities indicated that there existed a significant correlation between MSAP and S-SAP (r = 0.8118, 1,000 permutations, P < 0.05), whereas the correlation between MSAP and AFLP (r = 0.1048) is not statistically significant.




Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- S-SAP:
-
Sequence-specific amplification polymorphism
- MSAP:
-
Methylation-sensitive amplified polymorphism
References
Armstrong CL, Green CE (1985) Establishment and maintenance of friable, embryogenic maize callus and involvement of L-proline. Planta 164:207–214
Ashikawa I (2001) Surveying CpG methylation at 5′-CCGG in the genomes of rice cultivars. Plant Mol Biol 45:31–39
Bhaskaran S, Smith RA (1990) Regeneration in cereal tissue culture: a review. Crop Sci 30:1328–1336
Bohorova NE, Luna B, Briton RM, Huerta LD, Hoisington DA (1995) Regeneration potential of tropical, and subtropical, midaltitude, and highland maize inbreds. Maydica 40:275–281
Bregitzer P, Campbell RD, Wu Y (1995) Plant regeneration from barley callus: effects of 2,4-dichlorphenoxy acetic acid and phenylacetic acid. Plant Cell Tissue Organ Cult 43:229–235
Bregitzer P, Dahleen LS, Campbell RD (1998) Enhancement of plant regeneration from embryogenic callus of commercial barley cultivars. Plant Cell Rep 17:941–945
Carvalho CHS, Bohorova N, Bordallo PN, Abreu LL, Valicente FH, Bressan W, Paiva E (1997) Type II Callus production and plant regeneration in tropical maize genotypes. Plant Cell Rep 17:73–76
Cecchini E, Natali L, Cavallini A, Durante M (1992) DNA variations in regenerated plants of pea (Pisum sativum L.). Theor Appl Genet 84:874–879
Cervera MT, Ruiz-García L, Martínez-Zapater JM (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol Genet Genomics 268:543–552
Dahleen LS, Bregitzer P (2002) An improved media system for high regeneration rates from barley immature embryo-derived callus cultures of commercial cultivars. Crop Sci 42(3):934–938
Dong ZY, Wang YM, Zhang ZJ, Shen Y, Lin XY, Ou XF, Han FP, Liu B (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor Appl Genet 113:196–205
Duncan RP (1997) Tissue culture-induced variation and crop improvement. Adv Agron 58:201–240
Ellis THN, Poyser SJ, Knox MR, Vershinin AV, Ambrose MJ (1998) Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol Gen Genet 260:9–19
Etienne H, Bertrand B (2003) Somaclonal variation in Coffea arabica: effects of genotype and embryogenic cell suspension age on frequency and phenotype of variants. Tree Physiol 23:419–426
Golyasnaya NV, Tsvetkova NA (2006) Mismatch repair. Mol Biol 40:183–193
Gonzalgo ML, Jones PA (1997) Mutagenic and epigenetic effects of DNA methylation. Mutat Res 386:107–118
Guo JX, Jiang SC, Sun G (1998) Comparative study on remediation techniques of saline-alkali grassland in Songnen Plain. Chin J Appl Ecol 9:425–428
Guo WL, Wu R, Zhang YF, Liu XM, Wang HY, Gong L, Zhang ZH, Liu B (2007) Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth. et Hook. f. Plant Cell Rep. doi:10.1007/s00299-007-0320-0
Joyce SM, Cassells AC, Jain SM (2003) Stress and aberrant phenotypes in in vitro culture. Plant Cell Tissue Organ Cult 74(2):103–121
Kaeppler SM, Phillips RL (1993a) Tissue culture-induced DNA methylation variation in maize. Proc Natl Acad Sci USA 90:8773–8776
Kaeppler SM, Phillips RL (1993b) DNA methylation and tissue culture-induced variation in plants. In Vitro Cell Dev Biol 29:125–130
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188
Karp A (1995) Somaclonal variation as a tool for crop improvement. Euphytica 85:295–302
Kaup S, Grandjean V, Mukherjee R, Kapoor A, Keyes E, Seymour CB, Mothersill CE, Schofield PN (2006) Radiation-induced genomic instability is associated with DNA methylation changes in cultured human keratinocytes. Mutat Res 597:87–97
Kidwell KK, Osborn TC (1992) Simple plant DNA isolation procedures. In: Beckman JS, Osborn TC (eds) Plant genomes: methods for genetic and physical mapping. Kluwer Academic Publishers, Amsterdam, The Netherlands, pp 1–13
Larkin PJ, Scowcroft WR (1981) Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:443–455
Leigh F, Kalendar R, Lea V, Lee D, Donini P, Schulman AH (2003) Comparison of the utility of barley retrotransposon families for genetic analysis by molecular marker techniques. Mol Genet Genomics 269:464–474
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Manninen I, Schulman AH (1993) Bare-1, a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol Biol 22:829–846
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Matthes M, Singh R, Cheah SC, Karp A (2001) Variation in oil palm (Elais guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor Appl Genet 102:971–979
McClelland M, Nelson M, Raschke E (1994) Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res 22:3640–3659
Mohan JS (2001) Tissue culture-derived variation in crop improvement. Euphytica 118:153–166
Parra R, Pastor MT, Pérez-Payá E, Amo-Marco JB (2001) Effect of in vitro shoot multiplication and somatic embryogenesis on 5-methylcytosine content in DNA of Myrtus communis L. Plant Growth Regul 33:131–136
Peredo EL, Ángeles Revilla M, Arroyo-García R (2006) Assessment of genetic and epigenetic variation in hop plants regenerated from sequential subcultures of organogenic call. J Plant Physiol 163:1071–1079
Phillips RL, Kaeppler SM, Olhoft P (1994) Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc Natl Acad Sci USA 91:5222–5226
Porceddu A, Albertini E, Barcaccia G, Marconi G, Bertoli FB, Veronesi F (2002) Development of S-SAP markers based on an LTR-like sequence from Medicago sativa L. Mol Genet Genomics 267:107–114
Portis E, Acquadro A, Comino C, Lanteri S (2003) Analysis of DNA methylation during germination of pepper (Capsicum annuum L.) seeds using methylation-sensitive amplification polymorphism (MSAP). Plant Sci 166:169–178
Quemada H, Roth EK, Lark KG (1987) Changes in methylation status of tissue cultured soybean cells detected by digestion with the restriction enzymes Hpa II and Msp I. Plant Cell Rep 6:63–66
Rabinowicz PD, Citek R, Budiman MA, Nunberg A, Bedell JA, Lakey N, O’Shaughnessy AL, Nascimento LU, McCombie WR, Martienssen RA (2005) Differential methylation of genes and repeats in land plants. Genome Res 15:1431–1440
Reyna-López GE, Simpson J, Ruiz-Herrera J (1997) Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet 253:703–710
Rohlf FJ (2000) NTSYS-pc. Numerical taxonomy and multivariate analysis system. Ver. 2.1. Exeter software, Setauket, New York
Smulders MJM, Rus-Kortekaas W, Vosman B (1995) Tissue culture-induced DNA methylation polymorphisms in repetitive DNA of tomato calli and regenerated plants. Theor Appl Genet 91:1257–1264
Soleimani VD, Baum BR, Johnson DA (2005) Genetic diversity among barley cultivars assessed by sequence-specific amplification polymorphism. Theor Appl Genet 110:1290–1300
Soleimani VD, Baum BR, Johnson DA (2006) Quantification of the retrotransposon BARE-1 reveals the dynamic nature of the barley genome. Genome 49:389–396
Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M (2006) Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 9:199–207
Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, Grandbastien MA (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor Appl Genet 110:819–831
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang YM, Dong ZY, Zhang ZJ, Lin XY, Shen Y, Zhou D, Liu B (2005) Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170:1945–1956
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W (1997) Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253:687–694
Xiong LZ, Xu CG, Shagi-Maroof MA, Zhang Q (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261:439–446
Xu ML, Li XQ, Korban SS (2004) DNA-methylation alterations and exchanges during in vitro cellular differentiation in rose (Rosa hybrida L.). Theor Appl Genet 109:899–910
Acknowledgments
This study was supported by the Program for Changjiang Scholars and Innovative Research Team (PCSIRT) in University (#IRT0519) and the National Natural Science Foundation of China (30430060, 30471229). We are grateful to critical comments by two anonymous reviewers for improving the manuscript, and to Mr. Frederic Ngezahayo for editing the language.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaoling Li and Xiaoming Yu contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, X., Yu, X., Wang, N. et al. Genetic and epigenetic instabilities induced by tissue culture in wild barley (Hordeum brevisubulatum (Trin.) Link). Plant Cell Tiss Organ Cult 90, 153–168 (2007). https://doi.org/10.1007/s11240-007-9224-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-007-9224-5