Abstract
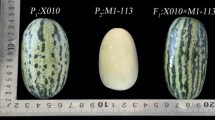
Plant architecture plays an important role in the yield, product quality, and cultivation practices of many crops. Branching pattern is one of the most important components in the plant architecture of melon (Cucumis melo L.). ‘Melon Chukanbohon Nou 4 Go’ (Nou-4) has a short-lateral-branching trait derived from a weedy melon, LB-1. This trait is reported to be controlled by a single recessive or incompletely dominant major gene called short lateral branching (slb). To find molecular markers for marker-assisted selection of this gene, we first constructed a linkage map using 94 F2 plants derived from a cross between Nou-4 and ‘Earl’s Favourite (Harukei-3)’, a cultivar with normal branching. We then conducted quantitative trait locus (QTL) analysis and identified two loci for short lateral branching. A major QTL in linkage group (LG) XI, at which the Nou-4 allele is associated with short lateral branching, explained 50.9 % of the phenotypic variance, with a LOD score of 12.5. We suggest that this QTL corresponds to slb because of the magnitude of its effect. Another minor QTL in LG III, at which the Harukei-3 allele is associated with short lateral branching, explained 9.9 % of the phenotypic variance, with a LOD score of 4.2. Using an independent population, we demonstrated that an SSR marker linked to the QTL in LG XI (slb) could be used to select for short lateral branching. This is the first report of mapping a gene regulating the plant architecture of melon.




Similar content being viewed by others
References
Aguilar-Martinez JA, Poza-Carrion C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19:458–472. doi:10.1105/tpc.106.048934
Brown A (1955) A mutant with suppressed lateral shoots. TGC Rep 5:6–7
Chiba N, Suwabe K, Nunome T, Hirai M (2003) Development of microsatellite markers in melon (Cucumis melo L.) and their application to major cucurbit crops. Breed Sci 53:21–27
Danin-Poleg Y, Reis N, Tzuri G, Katzir N (2001) Development and characterization of microsatellite markers in Cucumis. Theor Appl Genet 102:61–72
Denna D (1971) Expression of determinate habit in cucumbers. J Am Soc Hortic Sci 96:277–279
Diaz A, Fergany M, Formisano G, Ziarsolo P, Blanca J, Fei Z, Staub JE, Zalapa JE, Cuevas HE, Dace G, Oliver M, Boissot N, Dogimont C, Pitrat M, Hofstede R, van Koert P, Harel-Beja R, Tzuri G, Portnoy V, Cohen S, Schaffer A, Katzir N, Xu Y, Zhang H, Fukino N, Matsumoto S, Garcia-Mas J, Monforte AJ (2011) A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol 11:111. doi:10.1186/1471-2229-11-111
Doebley J, Stec A, Gustus C (1995) Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141:333–346
Dogimont C, Leconte L, Perin C, Thabuis A, Lecoq H, Pitrat M (2000) Identification of QTLs contributing to resistance to different strains of cucumber mosaic cucumovirus in melon. Acta Hortic 510:391–398
Essafi A, Díaz-Pendón JA, Moriones E, Monforte AJ, Garcia-Mas J, Martín-Hernández AM (2009) Dissection of the oligogenic resistance to Cucumber mosaic virus in the melon accession PI 161375. Theor Appl Genet 118:275–284. doi:10.1007/s00122-008-0897-x
Fan Z, Robbins MD, Staub JE (2006) Population development by phenotypic selection with subsequent marker-assisted selection for line extraction in cucumber (Cucumis sativus L.). Theor Appl Genet 112:843–855. doi:10.1007/s00122-005-0186-x
Fazio G, Staub JE, Chung SM (2002) Development and characterization of PCR markers in cucumber. J Am Soc Sci 127:545–557
Fazio G, Staub JE, Stevens MR (2003) Genetic mapping and QTL analysis of horticultural traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Theor Appl Genet 107:864–874. doi:10.1007/s00122-003-1277-1
Fernandez-Silva I, Eduardo I, Blanca J, Esteras C, Picó B, Nuez F, Arús P, Garcia-Mas J, Monforte AJ (2008) Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor Appl Genet 118:139–150. doi:10.1007/s00122-008-0883-3
Foster RE, Bond WT (1967) Abrachiate, an androecious mutant muskmelon. J Hered 58:13–14
Fukino N, Sakata Y, Kunihisa M, Matsumoto S (2007) Characterisation of novel simple sequence repeat (SSR) markers for melon (Cucumis melo L.) and their use for genotype identification. J Hortic Sci Biotechnol 82:330–334
Fukino N, Ohara T, Monforte AJ, Sugiyama M, Sakata Y, Kunihisa M, Matsumoto S (2008) Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.). Theor Appl Genet 118:165–175. doi:10.1007/s00122-008-0885-1
Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arús P, Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811. doi:10.1007/s00122-0041814-6
Harel-Beja R, Tzuri G, Portnoy V, Lotan-Pompan M, Lev S, Cohen S, Dai N, Yeselson L, Meir A, Libhaber SE, Avisar E, Melame T, Koert P, Verbakel H, Hofstede R, Volpin H, Oliver M, Fougedoire A, Stalh C, Fauve J, Copes B, Fei Z, Giovannoni J, Ori N, Lewinsohn E, Sherman A, Burger J, Tadmor Y, Schaffer AA, Katzir N (2010) A genetic map of melon highly enriched with fruit quality QTLs and EST markers, including sugar and carotenoid metabolism genes. Theor Appl Genet 121:511–533. doi:10.1007/s00122-010-1327-4
Hirabayashi T, Oizumi T, Sato K, Kote T, Yoshida S, Matsuo T, Komatsuka T (2007) Breeding of tendril-less type melon cultivar ‘TL Takami’ and its characteristics. Hort Res (Japan) 6:313–316 (Japanese with English summary)
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544. doi:10.1038/ng.591
Kaneko K, Miyagi M, Sakuma F (2006) Labor saving ability of training and fruit thinning of suppressed-branching melon in creeping cultivation. Bull Hortic Inst Ibaraki Agric Cent 14:9–14 (Japanese with English summary)
Kanno A, Uemura S (1980) Characteristics of the progeny derived from a cross between determinate type and normal type cultivar in cucumber. Annu Rep Morioka Branch Veg Ornam Crops Res Stn 5:31–33 (in Japanese)
Kobayashi T (1967) New cultivars of vegetables. 3. Pumpkin ‘Turunashi-kiji’. Bull Niigata Hortic Exp Stn 2:13–19
Kong Q, Xiang C, Yu Z (2006) Development of EST-SSRs in Cucumis sativus from sequence database. Mol Ecol Notes 6:1234–1236
Kosambi D (1943) The estimation of map distances from recombination values. Hum Genet 12:172–175
Kukita Y, Hayashi K (2002) Multicolor post-PCR labeling of DNA fragments with fluorescent ddNTPs. Biotechniques 33:502–506. doi:91200217
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newberg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li XZ, Yuan XJ, Jiang S, Pan JS, Deng SL, Wang G, Le He H, Wu AZ, Zhu LH, Koba T, Cai R (2008) Detecting QTLs for plant architecture traits in cucumber (Cucumis sativus L.). Breed Sci 58:453–460
Lin D, Wang T, Wang Y, Zhang X, Rhodes BB (1992) The effect of the branchless gene bl on plant morphology in watermelon. CGC Rep 15:74–75
Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15:31–39. doi:10.1016/j.tplants.2009.11.003
Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P (2011) Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J 67:701–714. doi:10.1111/j.1365-313X.2011.04629.x
Matsumoto S, Kunihisa M, Fukino N, Fujimura M (2005) Quick and easy DNA extraction from multiple samples of strawberry for classification of cultivars. Res Results Veg Tea Sci 2004:11–12 (in Japanese)
Minami M, Ujihara A, Campbell CG (1999) Morphology and inheritance of dwarfism in common buckwheat line, G410, and its stability under different growth conditions. Breed Sci 49:27–32
Monforte AJ, Oliver M, Gonzalo MJ, Alvarez JM, Dolcet-Sanjuan R, Arús P (2004) Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.). Theor Appl Genet 108:750–758. doi:10.1007/s00122-003-1483-x
Moreno E, Obando JM, Dos-Santos N, Fernández-Trujillo JP, Monforte AJ, Garcia-Mas J (2008) Candidate genes and QTLs for fruit ripening and softening in melon. Theor Appl Genet 116:589–602. doi:10.1007/s00122-007-0694-y
Napoli CA, Ruehle J (1996) New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J Hered 87:371–377
Ohara T, Wako T, Kojima A, Yoshida T, Ishiuchi D (2002) Breeding of suppressed-branching melon line ‘Melon chukanbohon nou 4’ (‘Melon parental line 4’) and its characteristics. Acta Hortic 588:227–231
Perchepied L, Bardin M, Dogimont C, Pitrat M (2005) Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology 95:556–565. doi:10.1094/phyto-95-0556
Rick CM (1980) Tomato linkage survey. TGC Rep 30:2–17
Ritschel PS, Lins TCDL, Tristan RL, Buso GSC, Buso JA, Ferreira ME (2004) Development of microsatellite markers from an enriched genomic library for genetic analysis of melon (Cucumis melo L.). BMC Plant Biol 4:9
Robbins MD, Casler M, Staub JE (2008) Pyramiding QTL for multiple lateral branching in cucumber using inbreed backcross lines. Mol Breed 22:131–139
Saeed M, Guo W, Ullah I, Tabbasam N, Zafar Y, Ur-Rahman M, Zhang T (2011) QTL mapping for physiology, yield and plant architecture traits in cotton (Gossypium hirsutum L.) grown under well-watered versus water-stress conditions. Electron J Biotechnol 14(3). doi:10.2225/vol14-issue3-fulltext-3
Sakata Y, Kubo N, Morishita M, Kitadani E, Sugiyama M, Hirai M (2005) QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus L.). Theor Appl Genet 112:243–250. doi:10.1007/s00122-005-0121-1
Sugiyama M, Ohara T, Sakata Y, Fukino N, Yoshioka Y, Shimomura K, Kojima A, Noguchi Y (2012) ‘Feria’, a new melon (Cucumis melo L.) cultivar with suppressed-branching and monoecious trait. Bull Natl Inst Veg Tea Sci 11:43–54 (in Japanese with English summary)
Takamure I, Kinoshita T (1985) Inheritance and expression of reduced culm number character in rice. Jpn J Breed 35:17–24
Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33:513–520
Umezaki T, Hiramatsu K (1997) Genetic characters of Akiyoshishirodaizu, a soybean dwarf variety. Jpn J Crop Sci 66:246–247
Wang S, Basten CJ, Zeng Z-B (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Yoshida T, Ishiuchi D (1994) Suppressed-branching in melon, characteristics and its inheritance. In: The 24th international horticultural congress, Kyoto, p 156
Yuste-Lisbona FJ, Capel C, Sarria E, Torreblanca R, Gómez-Guillamón ML, Capel J, Lozano R, López-Sesé AI (2010) Genetic linkage map of melon (Cucumis melo L.) and localization of a major QTL for powdery mildew resistance. Mol Breed 27:181–192. doi:10.1007/s11032-010-9421-5
Zalapa JE, Staub JE, McCreight JD, Chung SM, Cuevas H (2007) Detection of QTL for yield-related traits using recombinant inbred lines derived from exotic and elite US Western Shipping melon germplasm. Theor Appl Genet 114:1185–1201. doi:10.1007/s00122-007-0510-8
Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976
Zeng Z (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
We are grateful to Ms. H. Sainoki for her support in AFLP analysis. We also thank Ms. S. Negoro, T. Yamakawa, and S. Toyoda for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fukino, N., Ohara, T., Sugiyama, M. et al. Mapping of a gene that confers short lateral branching (slb) in melon (Cucumis melo L.). Euphytica 187, 133–143 (2012). https://doi.org/10.1007/s10681-012-0667-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0667-3