Abstract
Exploration into morphofunctional organisation of centromere DNA sequences is important for understanding the mechanisms of kinetochore specification and assembly. In-depth epigenetic analysis of DNA fragments associated with centromeric nucleosome proteins has demonstrated unique features of centromere organisation in chicken karyotype: there are both mature centromeres, which comprise chromosome-specific homogeneous arrays of tandem repeats, and recently evolved primitive centromeres, which consist of non-tandemly organised DNA sequences. In this work, we describe the arrangement and transcriptional activity of chicken centromere repeats for Cen1, Cen2, Cen3, Cen4, Cen7, Cen8, and Cen11 and non-repetitive centromere sequences of chromosomes 5, 27, and Z using highly elongated lampbrush chromosomes, which are characteristic of the diplotene stage of oogenesis. The degree of chromatin packaging and fine spatial organisations of tandemly repetitive and non-tandemly repetitive centromeric sequences significantly differ at the lampbrush stage. Using DNA/RNA FISH, we have demonstrated that during the lampbrush stage, DNA sequences are transcribed within the centromere regions of chromosomes that lack centromere-specific tandem repeats. In contrast, chromosome-specific centromeric repeats Cen1, Cen2, Cen3, Cen4, Cen7, Cen8, and Cen11 do not demonstrate any transcriptional activity during the lampbrush stage. In addition, we found that CNM repeat cluster localises adjacent to non-repetitive centromeric sequences in chicken microchromosome 27 indicating that centromere region in this chromosome is repeat-rich. Cross-species FISH allowed localisation of the sequences homologous to centromeric DNA of chicken chromosomes 5 and 27 in centromere regions of quail orthologous chromosomes.
Similar content being viewed by others
Introduction
In vertebrates, a centromere was first defined as a primary constriction visible in condensed chromosomes and later was described as a region of chromosome with a lower degree of recombination (Sumner 2003). Centromere regions of chromosomes are required for the attachment of mitotic and meiotic spindle, sister chromatid cohesion and regulation of chromosome segregation during cell division. Moreover, centromeres have several other important cell functions being involved in spatial genome organisation, heterochromatin assembly and chromocenter formation as well as regulation of expression for certain genes (Choo 2001; Watanabe 2004; Vos et al. 2006; Vagnarelli et al. 2008).
Formation of heterochromatin that often consists of highly repetitive satellite DNA is typical for centromere regions of chromosomes in a wide range of species (Hayakawa et al. 2003). After discovery of the phenomenon of RNA interference, it became evident that non-coding transcripts of centromere and pericentromere DNA are required for proper centromere functioning (Mellone and Allshire 2003; Almedia and Allshire 2005; Prasanth and Spector 2007; Hall et al. 2012). The importance of these highly specialised regions of chromosomes marked progress in our understanding how centromeres function in somatic and germ cells (Hayden and Willard 2012). At the same time, morphological organisation, epigenetic contents, degree of compaction and transcriptional activity of centromere chromatin require in-depth analysis.
To understand the mechanisms of kinetochore specification and assembly, it is important to explore the morphofunctional organisation of centromere DNA sequences. In a centromere region of chromosome, tandem or satellite repeats are currently subdivided into centromeric and pericentromeric repeats organised into head to tail arrays. Centromeric tandem repeats lie at the base of kinetochore and recruit the histone H3 variant CENP-A forming centromeric nucleosomes (Choo 2001). Notably, neocentromeres and immature centromeres that are devoid of satellite repeats usually consist of non-repetitive DNA that interacts with CENP-A (Burrack and Berman 2012). Thus, binding to CENP-A shown in biochemical studies (i.e. chromatin immunoprecipitation (ChIP), ChIP on chip technique, whole-genome ChIP sequencing and others) is a general feature of sequences responsible for centromere formation.
Among vertebrates, the domestic chicken (Gallus gallus domesticus) represents an adequate model for studying the difference between recently evolved centromeres and mature centromeres (Hori and Fukagawa 2012; Kalitsis and Choo 2012). Genome-wide characterisation of DNA fragments associated with CENP-A has demonstrated unique features of centromere organisation in chicken karyotype: there are both mature centromeres, comprising chromosome-specific tandem repeats, and recently evolved primitive centromeres, consisting of non-repetitive DNA sequences (Shang et al. 2010). In G. g. domesticus, centromere regions of the majority of gene-poor macrochromosomes (GGA1, GGA2, GGA3, GGA4, GGA7, GGA8) and microchromosome GGA11 bear non-identical centromere-specific repeats with different sizes of repetitive unit containing fragments of CR1 retrotransposon (chicken LINE). On the other hand, centromere regions of GGA5 and GGAZ have not accumulated satellite DNA and comprise unique DNA sequences interacting with CENP-A and enriched in retrotransposons (Shang et al. 2010). Similar to centromeric repeats, these unique DNA sequences that span only 30–50 kb have a clear centromeric function in kinetochore formation and chromosome segregation. For example, deletion of CenZ centromere DNA led to non-appearance of a kinetochore on chromosome Z in chicken DT40 cells (Shang et al. 2010). Other macrochromosomes, namely GGA6 and GGA9, and most of the gene-rich microchromosomes, typical for Galliform karyotypes, contain an identical 41 bp-repeat (CNM repeat) within their centromere regions (Matzke et al. 1990). CNM repeat is not centromere-specific and in addition to centromere regions localises in subtelomeric and interstitial regions of certain chicken chromosomes (Krasikova et al. 2006). The centromere region of one of the small microchromosomes, GGA27, similar to centromeres of GGA5 and GGAZ, is built by non-tandemly repetitive DNA sequences (Shang et al. 2010).
In this work, we analysed the precise distribution and transcriptional activity of recently identified chicken centromeric sequences using giant lampbrush chromosomes, which are characteristic of the diplotene stage of oogenesis. Meiotic lampbrush chromosomes (half-bivalents united by chiasmata) consist of arrays of chromomeres with lateral loops representing sites of the most intensive transcription (reviewed in Morgan 2002; Gaginskaya et al. 2009). The morphology of amphibian lampbrush centromeres from many previous investigations was originally systematised (Callan 1986; Macgregor 1986). It can be concluded that in avian and amphibian lampbrush chromosomes, centromeres have three major domains: compact loop-less chromomeres, one or several pairs of long lateral loops, and spherical protein bodies or granules that form at the primary constriction (Callan 1986; Krasikova and Gaginskaya 2010; Macgregor 2012).
Taking into account the postulated domain organisation of centromeres (Craig et al. 1999; Choo 2000, 2001), we have discriminated three functional domains in the centromere regions of lampbrush chromosomes: pairing domain (pricentromere heterochromatin), central domain (centromere chromatin), and pre-kinetochore region (Krasikova and Gaginskaia 2010). According to data reported by Edwards and Murray (2005), CENP-A protein is not concentrated at centromeres of Xenopus lampbrush chromosomes, and the position of a centromere can be defined by presence of centromeric repeats that are known to interact with CENP-A at other stages of the cell cycle and in other tissues. Importantly, in both avian and amphibian lampbrush chromosomes, pericentromeric compact chromomeres usually bear pairs of lateral loops, often very long, where RNA-polymerase II transcribes fragments of satellite DNA arrays (Diaz et al. 1981; Baldwin and Macgregor 1985; Barsacchi-Pilone et al. 1986; Solovei et al. 1996; Krasikova et al. 2006; Deryusheva et al. 2007). At the same time, transcriptional activity of truly centromeric sequences has not been studied in detail.
A remarkable round discrete object that was called centromere “protein body” forms at the primary constriction of all avian lampbrush centromeres (Gaginskaia et al. 1972; Solovei et al. 1996; Saifitdinova et al. 2003; Krasikova et al. 2006). Centromere protein bodies of chicken, quail and turkey lampbrush chromosomes are very small (approximately 1 μm in diameter) and look like centromere granules of amphibian lampbrush chromosomes (Krasikova et al. 2006, 2009; Macgregor 2012). A centromere protein body can be identified by marker components such as subunits of cohesin complex and component of synaptonemal lateral element SYCP3 (Krasikova et al. 2005). However, the accurate location and specific functions of centromere protein bodies in a centromere region of avian lampbrush chromosomes are still puzzling (Gaginskaya et al. 2009; Krasikova and Gaginskaia 2010).
Here, the arrangement of recently identified chicken centromeric sequences, which associate with CENP-A and include both repetitive and non-repetitive sequences, as well as their transcriptional activity were studied in detail. Using giant lampbrush chromosomes, we analysed the precise distribution of chicken centromeric repeats Cen1, Cen2, Cen3, Cen4, Cen7, Cen8, and Cen11 as well as recently evolved centromere-specific sequences of chromosomes 5, Z and 27 at high cytogenetic resolution.
Materials and methods
Chicken (G. g. domesticus) and Japanese quail (Coturnix coturnix japonica) lampbrush chromosomes were manually isolated from oocyte nuclei according to standard protocols (http://projects.exeter.ac.uk/lampbrush/protocols.htm). Preparations of lampbrush chromosomes were either dehydrated in 96 % ethanol and air-dried before fluorescent in situ hybridisation (FISH) or kept in 70 % ethanol before immunostaining.
Detection of centromere granules on lampbrush chromosomes was carried out by immunofluorescent staining with rabbit polyclonal antibodies K828 or K854 against STAG2 and Rad21 cohesin subunits, respectively, as previously described (Krasikova et al. 2006). Briefly, preparations were hydrated in 70–50–30 % ethanol, soaked in 1× phosphate buffered saline (PBS) with 0.02 % Tween-20 (Sigma) and incubated with a blocking reagent (Boehringer-Mannheim). Immunostaining with primary antibodies was followed by incubation with secondary antibodies conjugated with Cy3 (Amersham Pharmacia Biotech). After each incubation step, the preparations were washed in three changes of 1× PBS with 0.02 % Tween-20. After image acquisition, slides were washed in 4× saline-sodium citrate buffer (SSC: 3 M NaCl, 0.3 M sodium citrate, pH 7) with 0.1 % Tween-20 at 42 °C, dehydrated in 96 % ethanol, air-dried and used for FISH.
Chicken CENP-A associating sequences were mapped to lampbrush chromosomes by FISH with specific probes. Centromeric tandemly repetitive sequences specific for chicken chromosomes 1, 2, 3, 4, 7, 8, and 11 were amplified from chicken genomic DNA and labeled with biotin-16-dUTP by polymerase chain reaction (PCR) under standard conditions. Primers were designed according to corresponding reference sequences (Shang et al. 2010; accession numbers in DDBJ/EMBL/GenBank DNA database: AB556722 (Cen1), AB556723 (Cen2), AB556724 (Cen3), AB556725 (Cen4), AB556726 (Cen7), AB556728 (Cen11)) and are listed in “Supplementary Materials”. Centromeric tandem repeat Cen8 from GGA8 corresponds to the previously characterised partially inverted repeat (PIR) (Wang et al. 2002) and was amplified from chicken genomic DNA as described earlier (Krasikova et al. 2006).
DNA of BAC clones 231H07, 134D03, 066B03 from Female White Leghorn chicken BAC library (Hori et al. 2000), containing CENP-A associating non-repetitive sequences specific for chromosomes 5, Z and 27, respectively (Shang et al. 2010), was amplified and labeled with biotin-16-dUTP or digoxigenin-11-dUTP by degenerate oligonucleotide-primed PCR (DOP-PCR) with a degenerate 6-MW primer (Telenius et al. 1992). Additionally, some BAC clones from the Wageningen chicken BAC library (Crooijmans et al. 2000, http://www.bioinformatics.nl/gbrowse/cgi-bin/gbrowse) were used to analyse the transcriptional status of centromeric sequences at the lampbrush stage, to identify the chicken microchromosome 27 and to control the efficiency of Zoo-FISH. The full list of BAC clones used in this study is presented in Table S1. Labeled probes were dissolved in a standard hybridisation buffer (50 % formamide, 2× SSC, 10 % dextran sulphate) to a final concentration of 20–40 ng/μl with a 50-fold excess of salmon sperm DNA.
Chicken CNM repeat (Matzke et al. 1990; NCBI accession number X51431) and Japanese quail BglII-repeat (Tanaka et al. 2000; NCBI accession numbers AB035968–AB035970) were mapped to lampbrush chromosomes by FISH with specific oligonucleotide probes as described previously (Krasikova et al. 2006; Deryusheva et al. 2007).
For physical mapping of DNA sequences to lampbrush chromosomes, FISH was carried out according to a DNA/(DNA + RNA) hybridisation protocol (Krasikova et al. 2006; Deryusheva et al. 2007). Lampbrush chromosomes and probes were denatured together on a slide covered with a coverslip at 82 °C for 5 min. To investigate the transcriptional activity of centromere sequences at the lampbrush stage, a DNA/RNA hybridisation protocol was applied with only DNA probes being denatured. Hybridisation was performed at 37 °C or at room temperature (in case of FISH with oligonucleotides) in a humid chamber for 16–20 h. After hybridisation with long probes, the slides were washed in two changes of 0.2× SSC and two changes of 2× SSC at 60 °C. After FISH with oligonucleotides, the slides were washed in three changes of 2× SSC at room temperature. Detection of biotin-labeled and digoxigenin-labeled probes was carried out as described elsewhere (Galkina et al. 2006; Krasikova et al. 2006). To co-localise BACs and short tandem repeats on the same lampbrush chromosomes, two sets of differentially labeled probes were applied to the same slide sequentially. All preparations were dehydrated, air-dried and mounted in antifade solution containing 1 μg/ml DAPI.
Preparations were examined with a Leica fluorescence microscope DM4000 (100× objective) equipped with a monochrome digital camera DFC350 FX and appropriate filter cubes. CW 4000 FISH software was used to acquire fluorescent and phase contrast images that were further processed with Photoshop (Adobe Systems).
Results
High-resolution FISH mapping of chicken CENP-A—associating DNA sequences on giant lampbrush chromosomes
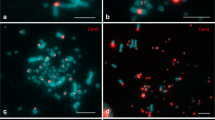
For the recently identified Cen1, Cen2, Cen3, Cen4, Cen7, Cen8 and Cen11 repeats (Shang et al. 2010), fragments that do not contain CR1 retrotransposon sequences have been amplified from the total chicken genomic DNA. FISH mapping data with the biotin-labeled probes on metaphase plates from chicken MDCC-MSB1 cells confirmed the amplified fragments to be centromere-specific (Fig. 1a). In the corresponding chicken lampbrush chromosomes, each of the Cen1, Cen2, Cen3, Cen4, Cen7, Cen8 and Cen11 centromeric repeats occupies one pair of chromomeres that were located within the condensed region of chromatin called ‘centromere bar’ (Fig. 1b, 2a, c, d, f, g). Centromeric repeats on macrochromosomes GGA1, GGA2, GGA3, GGA4, GGA7, GGA8 and the largest microchromosome GGA11 lie adjacent to the primary constriction and centromere granule revealed by immunofluorescent staining with antibodies against cohesin-complex proteins (Fig. 1c, 2b, b′, c, e, e′, f, g). Thus, chicken centromere-specific repeats are organised in two satellite arrays that are spatially presented by two distinct chromosomal domains or chromomeres. However, it is likely that only small portion of arrays of chromosome-specific tandem repeats is responsible for the formation of a centromere granule.
a Fluorescent in situ hybridisation (FISH) with a fragment of centromeric tandem Cen1 repeat (red) to chicken (G. g. domesticus) metaphase spreads. Arrows point to centromere region of chicken chromosome 1 (GGA1). b High-resolution FISH mapping of Cen1 repeat (red) on corresponding chicken lampbrush chromosome 1. c Fragment of lampbrush chromosome 1. FISH with Cen1 repeat (red) performed after immunofluorescent detection of centromere granules with antibodies against STAG2 (green). Chromosomes are counterstained with DAPI (blue). b′, c′ Corresponding phase contrast images. Arrows point to centromere regions on lampbrush chromosomes. Scale bar 10 μm. d, e Major sites of Cen1 localisation plotted on cytological chromomere-loop map of lampbrush chromosome 1. DAPI-positive chromomeres are represented by black or grey axial dots (Galkina et al. 2006). Chromomeres comprising Cen1 repeat are colored red. Green circle indicates centromere cohesin-enriched granule. Landmark structures are depicted: PBL11, marker loop; TBL, telomere bow-like loop
High-resolution FISH mapping of centromeric tandem repeats Cen2 (a, b, b′), Cen3 (c), Cen4 (d, e, e′), Cen7 (f) and Cen11 (g) (green) on chicken lampbrush chromosomes 2, 3, 4, 7 and 11, respectively. Centromere granules are revealed by immunofluorescent staining with antibodies against Rad21 or STAG2 (red). c, g Inserts show enlarged centromeric regions with fluorescent signals. b, e Enlarged view of GGA2 and GGA4 chromosomal regions around centromere. b′, e′ Corresponding phase contrast images. Chromosomes are counterstained with DAPI (blue). Arrows point to centromere regions on lampbrush chromosomes. Chromosome-specific centromeric tandem repeats occupy one or two DAPI-positive chromomeres surrounding the centromere granule. Scale bars 10 μm
Centromeric Cen1 repeat often hybridises not only to the GGA1 centromere but also to the telomere regions of all chromosomes as well as to the sites that correspond to the internal TTAGGG-repeat sites (Fig. 1). As we found, this hybridisation pattern can be explained by the presence of ‘CCCTAA’-sequence within the repetitive unit of centromeric Cen1 repeat. According to these data, appearance of interstitial TTAGGG site at GGA1 centromere reported earlier (Rodionov et al. 2002) is due to the presence of Cen1 repeat but not due to the fusion of ancestral chromosomes.
Surprisingly, recently evolved centromeric sequences that are responsible for kinetochore formation on chicken chromosomes 5 and Z demonstrate different spatial organisation during the lampbrush stage. In highly decondensed lampbrush chromosomes, CENP-A binding sequences Cen5 and CenZ cloned in BACs 231H07 and 134D03, respectively, were localised on the lateral loops that emerge from chromomeres adjacent to a centromere granule (Fig. 3a, b). We have also noticed that under standard conditions, DNA fragment contained in the BAC clone 231H07 hybridises with blocks of Z-macro-satellite (Fig. 3a) that is distributed on chicken chromosomes GGAZ, GGA1 and GGA3 and q-terminal region of GGA5 (Hori et al. 1996). This can be explained by enrichment of Cen5 sequence with specific transposable elements that have been amplified within blocks of Z-macro-satellite. An additional ВАС clone 429G11 next to non-centromeric satellite DNA on lampbrush chromosome 5 was mapped to lateral loops arising from a chromomere at a distance of 2 chromomeres from the centromere that corresponds approximately to 2–3 Mb (Fig. 3a). This result at high cytogenetic resolution confirms that in chicken chromosome 5 cluster of satellite repeat does not localise within a centromere region.
High-resolution FISH mapping of non-tandemly repetitive centromere-specific sequences on chicken lampbrush chromosomes. a Double FISH with BAC clone 231H07 containing CENP-A-binding centromeric sequences of GGA5 (green) and ВАС clone 429G11 containing sequences lying next to non-centromeric satellite DNA (red) to chicken lampbrush chromosome 5. b FISH with BAC clone 134D03 containing CENP-A-binding centromeric sequences of GGAZ (green) to chicken lampbrush chromosome Z. Insert shows enlarged centromeric region of another lampbrush chromosome Z with FISH signal. a′, b′ Corresponding phase contrast images. c FISH with BAC clone 066B03 containing CENP-A binding region of GGA27 (green) after immunofluorescent detection of centromere granule with antibodies against Rad21 (red). d FISH with centromeric BAC clone 066B03 (green) followed by ReFISH with CNM-repeat probe (yellow). Identification of chicken lampbrush microchromosome 27 by FISH with BAC clone WAG28K9 (red). All FISH signals are superimposed to phase contrast micrograph of lampbrush microchromosome 27. In lampbrush chromosome 27, CNM repeat cluster localises adjacent to non-repetitive centromeric sequences. FISH was performed according to DNA/(DNA + RNA) hybridisation protocol. Chromosomes are counterstained with DAPI (blue). Arrows point to centromere regions on lampbrush chromosomes. Scale bars 10 μm
In chicken lampbrush microchromosome 27, which is acrocentric, a BAC clone 066B03 containing CENP-A binding region localises in one pair of tiny lateral loops arising from prominent DAPI-positive chromomere within a centromere region (Fig. 3c, d). Note that the centromere granule detectable by antibodies against cohesin subunit Rad21 sits adjacent to this chromomere (Fig. 3c). Lampbrush microchromosome 27 was additionally identified by FISH with a BAC clone WAG28K9 from the Wageningen chicken BAC library (Fig. 3d). Multicolor FISH to chicken lampbrush chromosomes has demonstrated that prominent CNM repeat cluster (∼1–2 Mb) occupies chromomere adjacent to non-repetitive centromeric sequences in GGA27 (Fig. 3d).
Localisation of chicken non-repetitive centromere sequences on Japanese quail lampbrush chromosomes
Chicken CENP-A-associating tandem repeats from chromosomes 1, 2, 3, 4, 7, 8 and 11 do not hybridise to metaphase and lampbrush chromosomes of a closely related species—Japanese quail (C. c. japonica). As shown on example of chicken centromeric Cen2 repeat, these repeats are not detectable in quail karyotype by cross-species FISH (Zoo-FISH) (Fig. 4a, a′, in contrast to Fig. 2a). For an internal control of successful FISH, we added to hybridisation mix BAC clones from the Wageningen chicken BAC library that contain fragments of corresponding chicken chromosomes (Fig. 4; Table S1). Furthermore, centromeric chromosome-specific Cen1, Cen2, Cen3, Cen4, Cen7 and Cen11 repeats characteristic for chicken genome cannot be amplified from quail genomic DNA by PCR under standard conditions (data not shown) with the primers used (see Supplementary Information).
High-resolution localisation of chicken centromeric sequences on Japanese quail (C. c. japonica) lampbrush chromosomes. a, a′ Cross-species FISH with chicken centromeric tandem repeat Cen2 (green) and BAC clone WAG29F23 (red) to corresponding quail lampbrush chromosome 2 (CCO2). No signals can be detected on quail lampbrush chromosome 2 after FISH with Cen2 repeat. b, b′ Cross-species FISH with BAC clone 231H07 containing CENP-A-binding centromeric sequences of GGA5 (green) and BAC clone 429G11 containing sequences around GGA5-specific non-centromeric satellite repeat (red) to corresponding quail lampbrush chromosome 5 (CCO5). Bright signals from both BAC probes are visible at the centromere region of quail lampbrush chromosome 5. Chromosomes are counterstained with DAPI (blue). c Identification of quail ortholog of chicken microchromosome 27 by cross-species FISH with BAC clone WAG28K9 (red) and localisation of sequences homologous to CENP-A binding region of GGA27 (BAC clone 066B03, green). c′ ReFISH with BglII-repeat probe (yellow) on the same lampbrush microchromosome. c″ All three signals can be easily resolved on the merged image of this tiny lampbrush microchromosome. a′, b′, c-c″ Fluorescent images are superimposed to phase contrast micrographs. Arrows point to centromere regions on lampbrush chromosomes. FISH was performed according to DNA/(DNA + RNA) hybridisation protocol. Scale bars 10 μm
A different situation was observed for non-repetitive centromeric sequences of chicken chromosomes 5 and 27. By means of Zoo-FISH, we have demonstrated the presence of sequences homologous to chicken Cen5 DNA in a centromere region of Japanese quail chromosome 5 (CCO5) without changes in the chromosomal position of the fragment. In quail lampbrush chromosome 5, BAC clone 231H07 containing chicken Cen5 DNA hybridised to a single transcription unit that emerged from enlarged DAPI-positive chromomere (Fig. 4b, b′). This particular chromomere was previously shown to be associated with cohesin-enriched centromere granule in quail lampbrush chromosome 5 (Daks et al. 2010). A BAC clone 429G11 containing sequences around GGA5-specific non-centromeric satellite repeat also efficiently hybridised to lateral loops extended from chromomere next to the prominent centromeric one (Fig. 4b, b′).
Japanese quail ortholog of chicken microchromosome 27 in the lampbrush form was identified by cross-species FISH with BAC WAG28K9 (Fig. 4c, c″). Sequences homologous to chicken Cen27 (BAC clone 066B03) localise in a centromere region of the orthologous quail microchromosome (Fig. 4c, c″), which is submetacentric presumably due to accumulation of heterochromatin on its short arm (Krasikova et al. 2009). In the centromere region of Japanese quail ortholog of chicken microchromosome 27, non-tandemly repetitive sequences lie adjacent to a prominent array of BglII-repeat (Fig. 4c′, c″) that belongs to a 41-bp tandem repeat family (Tanaka et al. 2000). However, we have not detected any signals after Zoo-FISH of chicken centromeric DNA fragment containing CenZ on quail lampbrush chromosome Z (data not shown). This could be due to a high degree of divergence of this sequence which resulted in a non-effective hybridisation.
Transcriptional activity of DNA sequences within the centromere regions of chicken lampbrush chromosomes
Transcriptional activity of the GGA1, GGA2, GGA3, GGA4, GGA7, GGA8 and GGA11 centromeric repeats during the lampbrush stage of oogenesis was examined by FISH according to DNA/RNA hybridisation protocol. In contrast to 41-bp repeats (CNM and PO41), centromere clusters of the Cen1, Cen2, Cen3, Cen4, Cen7, Cen8 and Cen11 repeats do not demonstrate any transcriptional activity on chicken lampbrush chromosomes. These chromosome-specific centromeric repeats do not hybridise to the lateral loops, where active transcription takes place, of their corresponding lampbrush chromosomes (Figs. 1 and 2). In addition, no signal can be detected on corresponding lampbrush chromosomes if chromosomal DNA denaturation is omitted before FISH with the labeled fragments (Fig. 5b, d). In these experiments, BAC clones from the Wageningen chicken BAC library that comprise fragments transcribed during the lampbrush stage (Zlotina et al. 2012) were used as a positive control to verify the integrity of RNA on lampbrush chromosome preparations (Table S1).
Examples of transcriptional activity analysis of chicken centromeric tandem repeats during the lampbrush stage. a, a′, b Double FISH with centromeric repeat Cen3 (green) and BAC clone WAG13D11 (red) on chicken lampbrush chromosome 3. c, d Double FISH with centromeric repeat Cen11 (green) and BAC clone WAG12F3 on chicken lampbrush chromosome 11. FISH was performed according to DNA/(DNA + RNA) (a, a′, c) or DNA/RNA (b, d) hybridisation protocols. a Insert shows enlarged centromeric region with fluorescent signals. a′, c Fluorescent images are superimposed to phase contrast micrographs. No signals from probes to centromeric repeats can be detected if chromosomal DNA denaturation is omitted. Chromosomes are counterstained with DAPI (blue). Arrows point to centromere regions on lampbrush chromosomes. Scale bars 10 μm
For example, in case of chicken lampbrush chromosome 3, BAC clone WAG13D11 hybridised to the lateral loops in both DNA/(DNA + RNA) and DNA/RNA FISH experiments, while centromeric Cen3 repeat hybridised only in case of prior denaturation of chromosomal DNA (Fig. 5a, a′, b). Similarly, FISH with centromeric Cen11 repeat gave prominent fluorescent signals on lampbrush chromosome 11 only when FISH was performed according to DNA/(DNA + RNA) hybridisation protocol. At the same time, BAC probe WAG12F3 added to the same hybridisation mix gave bright labeling of small lateral loops near the centromere region even without DNA denaturation indicating its hybridisation to nascent transcripts (Fig. 5c, d).
Surprisingly, BAC clones 134D03, 231H07 and 066B03 containing centromeric sequences of chromosomes GGAZ, GGA5 and GGA27, respectively, hybridise to transcription units in chicken lampbrush centromeres (Fig. 3). These BAC clones contain not only sequences interacting with CENP-A, but also sequences of neighborhood regions (Shang et al. 2010).
Computer analysis of chicken centromere sequences
One of the recent studies reports presence of full-length L1 retrotransposon in human 10q25 neocentromere (Chueh et al. 2009). The authors demonstrated that this particular copy of L1 retrotransposon is transcriptionally active with transcripts being incorporated into CENP-A-associated chromatin and important for regulation the neocentromeric chromatin. Using RepeatMasker software (www.girinst.org), we have performed a search for functional copies of retrotransposons of class LINE (Long Interspersed Nuclear Element) and LTR (long terminal repeat)-containing repeats in CENP-A interacting sequences of chicken chromosomes 5, Z and 27 (accession numbers in DDBJ/EMBL/GenBank DNA database: AB556729–AB556731, Shang et al. 2010). All copies of CR1 (chicken LINE) interspersed within non-tandemly organised centromeric Cen5, CenZ and Cen27 sequences were found to be 5′-truncated and thus devoid of active promoters. Such distribution of CR1 interspersed repeat is in correspondence with modern data on distribution and functional activity of LINE class repeats in chicken genome, which contains around 26,000 copies of the CR1 element with the vast majority of copies being inactive (ICGSC 2004; Wicker et al. 2005; Lee et al. 2009).
Apart from CR1 fragments, multiple copies of LTR-retrotransposons were detected in CENP-A binding sequences specific for GGA5, GGAZ and GGA27 centromeres. In centromeric sequences of chromosomes Z and 27, we have found several copies of LTR elements with fully intact LTR containing promoter region at least at one of the ends. For example, at terminal regions of both centromeric sequences, there is a full-length copy of LTR retrotransposon Kronos (Gypsy family), as well as copies of endogenous retrovirus GGERVLA, belonging to one of the youngest group of transposons that diverged recently and thus retained their functional activity in the genome (ICGSC 2004; Huda et al. 2008).
Using computer analysis of chicken CENP-A binding sequences, we have demonstrated that chicken centromeric repeats Cen7, Cen8 and Cen11 have motifs similar to mammalian CENP-B box (Masumoto et al. 1989) (Supplementary information). Some of these motifs are longer then conservative CENP-B box alike CENP-B box in pigeon centromeric satellite repeat PR1 (Solovei et al. 1996). Previously, highly conserved DNA-binding protein CENP-B was identified in most chicken centromeres (Saffery et al. 1999). Thus, chicken centromeric repeats, at least those of chromosomes GGA7, GGA8 and GGA11, potentially have features that allow them to attract CENP-B protein.
Discussion
Many recent studies have focused on the identification and high-resolution mapping of sequences that were not characterised during the vertebrate genome sequencing projects including projects directed to sequencing avian genomes (ICGSC 2004). Although the centromere-specific variant of histone H3 (CENP-A) was not detected in avian and amphibian lampbrush chromosomes (Edwards and Murray 2005; Krasikova and Gaginskaia 2010), recently identified sequences binding to CENP-A in chicken somatic cells were successfully mapped at high resolution on chicken lampbrush chromosomes. At the lampbrush stage, CENP-A interacting tandem repeats and non-repetitive centromeric sequences in all cases were assigned adjacent to centromere granule enriched with cohesin subunits. These data for the first time directly indicate that centromere protein bodies or granules could serve as a universal and reliable marker for centromere mapping on lampbrush chromosomes. Moreover, we found that the degree of chromatin packaging and fine spatial organisation of tandem centromeric repeats and non-repetitive centromeric sequences differ significantly at the lampbrush stage of oogenesis when chromosomes acquire chromomere-loop form. However, in terms of protein body formation, all centromeres of chicken lampbrush chromosomes have similar morphology. It also appears that other loop-less chromomeres within the centromere bars of the GGA 1, 2, 3, 4, 7, 8 and 11 are occupied by sequences of chromosome arms but not by tandem repeats.
Importantly, the chicken genome has a three-hybrid centromere structure; in addition to centromeres organised as extended arrays of tandem repeats and centromeres consisting of interspersed repeats and unique DNA sequences (Shang et al. 2010), some chromosomes bear both tandem repeats and non-repetitive centromeric DNA. One of such examples is microchromosome 27, which as we found bears centromere with accumulated CNM repeats, possibly ensuring the stability of a microchromosomal centromere. Because GGA27 is a very small microchromosome (euchromatic part is approximately 5.2 Mb), the relative positions of unique and tandemly repetitive sequences were precisely determined only at the lampbrush stage. By ChIP sequencing, CNM repeat was previously shown to interact with CENP-A (Shang et al. 2010). Thus, centromere region of GGA27 is not satellite-free and consists of minimal unique Cen27 sequences and retrotransposons that bind to CENP-A, which are surrounded by one or two arrays of CNM repeat. The centromere gap in the sequence assembly of GGA27 should be thus filled with CNM repetitive sequence. Comparative mapping demonstrated that similar organisation of a centromere region is typical for quail orthologous microchromosome, in which non-tandemly repetitive sequences were localised in close proximity to an array of BglII-repeat. In chicken microchromosome 27 and quail orthologous microchromosome, 41 bp repeats could be involved into kinetochore formation or instead could serve as pericentromeric repeats.
It is still unknown whether unique centromere-specific sequences can also be typical for those chicken centromeres that consist of arrays of tandem repeats. If this is true, non-tandemly repetitive centromere regions could accumulate centromeric repeats and thus blocks of heterochromatin during the centromere growth period according to postulated centromere life cycle (Kalitsis and Choo 2012). On the other hand, centromeres of GGAZ and GGA5 cannot be surrounded by arrays of tandem repeats in pericentric regions because in the chicken genome assembly CenZ and Cen5 sequences interacting with CENP-A map to long contigs that are not interrupted by gaps.
We have also provided evidence that there is dissimilarity between domestic chicken (G. g. domesticus) and Japanese quail (C. c. japonica) centromeric tandem repeats at least in some macrochromosomes. It can be concluded that during divergence of these two species, chromosome-specific centromeric tandem repeats of chicken macrochromosomes evolved independently. On the contrary, sequences responsible for centromere formation on GGA5 and GGA27 are conserved between chicken and quail. Whether these sequences in Japanese quail genome have a similar function in kinetochore formation and interact with CENP-A remains an open question.
In somatic cells, transcripts of pericentromeric DNA sequences are involved in the formation of pericentromeric heterochromatin (Fukagawa et al. 2004; Buhler and Moazed 2007; Zaratiegui et al. 2011; Hall et al. 2012). Pericentromeric DNA repeat transcripts synthesised during the lampbrush stage may have a function in the formation of pericentromeric heterochromatin during the early stages of embryogenesis according to the mechanism of co-transcriptional gene silencing. We demonstrate here that in chicken oocytes, truly centromeric (CENP-A interacting) tandem repeats are not transcribed during the lampbrush stage of oogenesis. Likewise, chaffinch centromeric highly repeated sequence FCP is not transcribed on lampbrush chromosomes (Saifitdinova et al. 2003) and contains a region similar to CENP-B box (see Supplementary information). This is in accordance with our working hypothesis that states that pericentromeric but not centromeric repeats are transcribed during the lampbrush stage (Krasikova and Gaginskaya 2010).
In contrast, non-repetitive сentromeric sequences of GGAZ, GGA5 and GGA27 are transcribed on lateral loops of lampbrush chromosomes. Together with that, the centromere identity of these chromosomes is not lost during the lampbrush stage. It cannot be ruled out that in case of hybridisation of BAC clones containing these sequences to chicken lampbrush chromosomes, we have detected transcripts of not only true centromeric but also pericentromeric non-repetitive DNA sequences that are transcribed without boundary. Alternatively, natural transcription of unique non-repetitive centromeric sequences observed on chicken lampbrush chromosomes Z, 5 and 27 may be required for proper “centromerisation” (Hall et al. 2012). Because transcription units on avian lampbrush chromosomes are tightly covered by RNA polymerases (reviewed in Gaginskaya et al. 2009), the number of nucleosomes could be very low in CENP-A interacting regions of lampbrush chromosomes Z and 5.
Bioinformatic analysis allows to suggest that truncated LINE retrotransposable elements are not able to initiate the transcription of non-protein-coding centromere regions of chicken chromosomes 5, Z and 27. At the same time, the data presented here indirectly indicate that transcription of these centromeric regions during the lampbrush stage could be initiated by active promoters of LTR-containing retrotransposons or even solo-LTRs that lie adjacent to transcribing sequences. Similar data on appearance of active LTR promoters near clusters of chicken CNM and PO41 repeats that are known to be transcribed was reported previously (Deryusheva et al. 2007).
It can be assumed that FISH analysis and immunofluorescent assays with the usage of giant lampbrush chromosomes allow for a comprehensive investigation of functional and highly specialised regions of chromosomes such as centromeres.
Abbreviations
- Ab:
-
Antibody
- BAC:
-
Bacterial artificial chromosome
- CCO:
-
Japanese quail (Coturnix coturnix japonica) chromosome
- Cen:
-
Centromere-specific sequence
- CENP:
-
Centromere protein
- ChIP:
-
Chromatin immunoprecipitation
- CNM:
-
Chicken nuclear-membrane-associated repeat
- CR1:
-
Chicken Repetitive 1 Elements
- FISH:
-
Fluorescent in situ hybridisation
- GGA:
-
Chicken (Gallus gallus) chromosome
- LINE:
-
Long interspersed nuclear elements
- LTR:
-
Long terminal repeats
- Mb:
-
Megabase
- PBS:
-
Phosphate buffered saline
- PIR:
-
Partially inverted repeat
- SSC:
-
Saline-sodium citrate buffer
- SYCP3:
-
Synaptonemal complex protein 3
- Zoo-FISH:
-
Cross-species FISH
References
Almeida R, Allshire RC (2005) RNA silencing and genome regulation. Trends Cell Biol 15:251–258
Baldwin L, Macgregor HC (1985) Centromeric satellite DNA in the newt Triturus cristatus karelinii and related species: its distribution and transcription on lampbrush chromosomes. Chromosoma 92:100–107
Barsacchi-Pilone G, Batistoni R, Andronico F, Vitelli L, Nardi I (1986) Heterochromatic DNA in Triturus (Amphibia, Urodela). I. A satellite DNA component of the pericentric C-bands. Chromosoma 93:435–446
Bühler M, Moazed D (2007) Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14:1041–1048
Burrack LS, Berman J (2012) Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res 20:607–619
Callan HG (1986) Lampbrush chromosomes. Mol Biol Biochem Biophys 36:1–254
Choo KH (2000) Centromerization. Trends Cell Biol 10:182–188
Choo KH (2001) Domain organization at the centromere and neocentromere. Dev Cell 1:165–177
Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5:e1000354
Craig JM, Earnshaw WC, Vagnarelli P (1999) Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp Cell Res 246:249–262
Crooijmans RP, Vrebalov J, Dijkhof RJ, van der Poel JJ, Groenen MA (2000) Two-dimensional screening of the Wageningen chicken BAC library. Mamm Genome 11:360–363
Daks AA, Deriusheva SE, Krasikova AV, Zlotina AM, Gaginskaia ER, Galkina SA (2010) Lampbrush chromosomes of the Japanese quail (Coturnix coturnix japonica): a new version of cytogenetic maps. Genetika 46:1335–1338
Deryusheva S, Krasikova A, Kulikova T, Gaginskaya E (2007) Tandem 41-bp repeats in chicken and Japanese quail genomes: FISH mapping and transcription analysis on lampbrush chromosomes. Chromosoma 116:519–530
Diaz MO, Barsacchi-Pilone G, Mahon KA, Gall JG (1981) Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell 24:649–659
Edwards NS, Murray AW (2005) Identification of xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell 16:1800–1810
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Gaginskaya ER (1972) The nuclear structures in oocytes of adult birds. II. Protein bodies and the karyosphere. Tsitologiia 14:568–577
Gaginskaya E, Kulikova T, Krasikova A (2009) Avian lampbrush chromosomes: a powerful tool for exploration of genome expression. Cytogenet Genome Res 124:251–267
Galkina S, Deryusheva S, Fillon V, Vignal A, Crooijmans R, Groenen M, Rodionov A, Gaginskaya E (2006) FISH on avian lampbrush chromosomes produces higher resolution gene mapping. Genetica 128:241–251
Hall LE, Mitchell SE, O’Neill RJ (2012) Pericentric and centromeric transcription: a perfect balance required. Chromosome Res 20:535–546
Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y (2003) Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci 116:3327–3338
Hayden KE, Willard HF (2012) Composition and organization of active centromere sequences in complex genomes. BMC Genomics 13:324
Hori T, Fukagawa T (2012) Establishment of the vertebrate kinetochores. Chromosome Res 20:547–561
Hori T, Suzuki Y, Solovei I, Saitoh Y, Hutchison N, Ikeda JE, Macgregor H, Mizuno S (1996) Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res 4:411–426
Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S (2000) Wpkci, encoding an altered form of PKCI, is conserved widely on the avian W chromosome and expressed in early female embryos: implication of its role in female sex determination. Mol Biol Cell 11:3645–3660
Huda A, Polavarapu N, Jordan IK, McDonald JF (2008) Endogenous retroviruses of the chicken genome. Biol Direct 3:9
International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Kalitsis P, Choo KH (2012) The evolutionary life cycle of the resilient centromere. Chromosoma 121:327–340
Krasikova AV, Gaginskaia ER (2010) Organization of centromere regions of chromosomes in the lampbrush phase. Tsitologiia 52:515–533
Krasikova A, Barbero JL, Gaginskaya E (2005) Cohesion proteins are present in centromere protein bodies associated with avian lampbrush chromosomes. Chromosome Res 13:675–685
Krasikova A, Deryusheva S, Galkina S, Kurganova A, Evteev A, Gaginskaya E (2006) On the positions of centromeres in chicken lampbrush chromosomes. Chromosome Res 14:777–789
Krasikova A, Daks A, Zlotina A, Gaginskaya E (2009) Polymorphic heterochromatic segments in Japanese quail microchromosomes. Cytogenet Genome Res 126:148–155
Lee SH, Eldi P, Cho SY, Rangasamy D (2009) Control of chicken CR1 retrotransposons is independent of Dicer-mediated RNA interference pathway. BMC Biol 7:53
Macgregor HC (1986) The lampbrush chromosomes of animal oocytes. In: Risley MS (ed) Chromosome structure and function. Van Rostrand Remhold, New York, pp 152–186
Macgregor HC (2012) Chromomeres revisited. Chromosome Res 7: DOI: 10.1007/s10577-012-9310-3
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109:1963–1973
Matzke MA, Varga F, Berger H, Schernthaner J, Schweizer D, Mayr B, Matzke AJ (1990) A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma 99:131–137
Mellone BG, Allshire RC (2003) Stretching it: putting the CEN(P-A) in centromere. Curr Opin Genet Dev 13:191–198
Morgan GT (2002) Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res 10:177–200
Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev 21:11–42
Rodionov AV, Lukina NA, Galkina SA, Solovei I, Saccone S (2002) Crossing over in chicken oogenesis: cytological and chiasma-based genetic maps of the chicken lampbrush chromosome 1. J Hered 93:125–129
Saffery R, Earle E, Irvine DV, Kalitsis P, Choo KH (1999) Conservation of centromere protein in vertebrates. Chromosome Res 7:261–265
Saifitdinova A, Derjusheva S, Krasikova A, Gaginskaya E (2003) Lampbrush chromosomes of the chaffinch (Fringilla coelebs L.). Chromosome Res 11:99–113
Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T (2010) Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 20:1219–1228
Solovei IV, Joffe BI, Gaginskaya ER, Macgregor HC (1996) Transcription of lampbrush chromosomes of a centromerically localized highly repeated DNA in pigeon (Columba) relates to sequence arrangement. Chromosome Res 4:588–603
Sumner AT (2003) Chromosomes: organization and function. Blackwell Publishing, Oxford
Tanaka K, Suzuki T, Nojiri T, Yamagata T, Namikawa T, Matsuda Y (2000) Characterization and chromosomal distribution of a novel satellite DNA sequence of Japanese quail (Coturnix coturnix japonica). J Hered 91:412–415
Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725
Vagnarelli P, Ribeiro SA, Earnshaw WC (2008) Centromeres: old tales and new tools. FEBS Lett 582:1950–1959
Vos LJ, Famulski JK, Chan GK (2006) How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol 84:619–639
Wang X, Li J, Leung FC (2002) Partially inverted tandem repeat isolated from pericentric region of chicken chromosome 8. Chromosome Res 10:73–82
Watanabe Y (2004) Modifying sister chromatid cohesion for meiosis. J Cell Sci 117:4017–4023
Wicker T, Robertson JS, Schulze SR, Feltus FA, Magrini V, Morrison JA, Mardis ER, Wilson RK, Peterson DG, Paterson AH, Ivarie R (2005) The repetitive landscape of the chicken genome. Genome Res 15:126–136
Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, Cande WZ, Antequera F, Arcangioli B, Martienssen RA (2011) RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479:135–138
Zlotina A, Galkina S, Krasikova A, Crooijmans RPMA, Groenen MAM, Gaginskaya E, Deryusheva S (2012) Centromere positions in chicken and Japanese quail chromosomes: de novo centromere formation versus pericentric inversions. Chromosome Res (in press)
Acknowledgments
The authors gratefully acknowledge Richard Crooijmans and Martin Groenen (the Wageningen University) for providing BAC clones from the Wageningen chicken BAC library. This investigation was supported by Russian Foundation for Basic Research (Grant #11-04-01347-а) and Federal Grant-in-Aid Program «Human Capital for Science and Education in Innovative Russia». The authors acknowledge Saint-Petersburg State University for a research grant #1.38.66.2011 and the Core Facility ‘CHROMAS’ (SPbSU) for an access to equipment.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 17 kb)
Rights and permissions
About this article
Cite this article
Krasikova, A., Fukagawa, T. & Zlotina, A. High-resolution mapping and transcriptional activity analysis of chicken centromere sequences on giant lampbrush chromosomes. Chromosome Res 20, 995–1008 (2012). https://doi.org/10.1007/s10577-012-9321-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-012-9321-0