Abstract
The genus Tokudaia comprises three species, two of which have lost their Y chromosome and have an XO/XO sex chromosome constitution. Although Tokudaia muenninki (Okinawa spiny rat) retains the Y chromosome, both sex chromosomes are unusually large. We conducted a molecular cytogenetic analysis to characterize the sex chromosomes of T. muenninki. Using cross-species fluorescence in situ hybridization (Zoo-FISH), we found that both short arms of the T. muenninki sex chromosomes were painted by probes from mouse chromosomes 11 and 16. Comparative genomic hybridization analysis was unable to detect sex-specific regions in the sex chromosomes because both sex probes highlighted the large heterochromatic blocks on the Y chromosome as well as five autosomal pairs. We then performed comparative FISH mapping using 29 mouse complementary DNA (cDNA) clones of the 22 X-linked genes and the seven genes linked to mouse chromosome 11 (whose homologue had fused to the sex chromosomes), and FISH mapping using two T. muenninki cDNA clones of the Y-linked genes. This analysis revealed that the ancestral gene order on the long arm of the X chromosome and the centromeric region of the short arm of the Y chromosome were conserved. Whereas six of the mouse chromosome 11 genes were also mapped to Xp and Yp, in addition, one gene, CBX2, was also mapped to Xp, Yp, and chromosome 14 in T. muenninki. CBX2 is the candidate gene for the novel sex determination system in the two other species of Tokudaia, which lack a Y chromosome and SRY gene. Overall, these results indicated that the Y chromosome of T. muenninki avoided a loss event, which occurred in an ancestral lineage of T. osimensis and T. tokunoshimensis, through fusion with an autosome. Despite retaining the Y chromosome, sex determination in T. muenninki might not follow the usual mammalian pattern and deserves further investigation.
Similar content being viewed by others
Introduction
The genus Tokudaia belongs to the subfamily Murinae (Muridae, Rodentia) and consists of three species: Tokudaia muenninki (Okinawa spiny rat), Tokudaia osimensis (Amami spiny rat), and Tokudaia tokunoshimensis (Tokunoshima spiny rat). Each species is endemic to an island in the southernmost part of Japan and is named after the island on which it is found. The sex chromosomes in this genus are unusual among mammals. The diploid chromosome numbers of T. osimensis and T. tokunoshimensis are an odd number, 25 and 45, respectively. Neither T. osimensis nor T. tokunoshimensis has a Y chromosome and sex is determined with an XO/XO sex chromosome constitution in both species (Honda et al. 1977, 1978; Kobayashi et al. 2007). Although the diploid chromosome number of T. muenninki is 44 with XX/XY sex chromosome constitution, both sex chromosomes are unusually large (Tsuchiya et al. 1989; Murata et al. 2010). The euchromatic regions of the X and Y chromosomes occupy 8% and 4% of the haploid genome, respectively, which suggests that an autosomal addition has occurred (Murata et al. 2010). On the basis of molecular phylogenetic analysis, T. muenninki was the first species in the genus to diverge, which suggests that the Y chromosome was lost in the common ancestor of T. osimensis and T. tokunoshimensis after this event (Murata et al. 2010). Three stepwise events that led to the disappearance of the Y chromosome have been suggested; gene transpositions from the Y chromosome to autosomes, superseding Y-linked genes by new genes on the X chromosome or autosomes, and Y-to-X translocation containing Y-linked genes (Arakawa et al. 2002; Kuroiwa et al. 2010).
The SRY gene, which is the master gene for male sex determination in placental mammals (Sinclair et al. 1990; Koopman et al. 1991) and is located on the Y chromosome, has been lost completely in T. osimensis and T. tokunoshimensis (Soullier et al. 1998; Sutou et al. 2001). Other Y chromosome genes have also been lost in T. osimensis, including RBMY1A1 (RNA binding motif protein, Y chromosome, family 1, member A1) (Kuroiwa et al. 2010), which plays a role in spermatogenesis (Mazeyrat et al. 1999; Elliott 2004). However, most proto-Y-linked genes have been conserved in both sexes of T. osimensis and T. tokunoshimensis through translocation from the Y chromosome to the distal region of the long arm of the X chromosome. The absence of SRY indicates that T. osimensis and T. tokunoshimensis must have a novel sex-determining mechanism. Recently, CBX2 (chromobox homolog 2, synonym is M33) has emerged as a candidate gene for sex determination in these species (Kuroiwa et al. 2011). CBX2 is a single copy gene that is located on an autosome in mice and humans. However, quantification of the copy number of CBX2 in T. osimensis and T. tokunoshimensis by quantitative real-time PCR showed that there were two or three more copies of CBX2 per haploid genome in males than in females, which suggests that CBX2 might be involved in a novel sex-determining mechanism through gene dosage (Kuroiwa et al. 2011).
Although T. muenninki possesses a Y chromosome, several aspects of sex determination in this species are unusual (Tsuchiya et al. 1989; Murata et al. 2010). Multiple copies of the SRY gene (>70) are found in T. muenninki, and these are distributed along the entire long arm of the Y chromosome (Murata et al. 2010). Most copies are not functional; however, an open reading frame (ORF) is conserved in three copies (Murata et al. 2010). A single amino acid substitution that is specific to T. muenninki was detected in the DNA binding surface domain in the high-mobility group (HMG)-box of all SRY copies (Murata et al. 2010). This region is important for the binding of SRY to target DNA sequences, which suggests that the DNA-binding ability of all SRY proteins produced in T. muenninki is weakened by this mutation.
To reveal the composition of the X and Y chromosomes in T. muenninki, we performed cross-species fluorescence in situ hybridization (Zoo-FISH) with chromosome-specific DNA probes constructed from the laboratory mouse (Mus musculus). To describe the composition in more detail and identify sex-specific regions in the sex chromosomes, we performed comparative genomic hybridization (CGH) analysis. When this provided limited information, we then conducted comparative FISH analysis using 29 mouse cDNA clones of functional genes linked to the X chromosome and chromosome 11 and FISH mapping using two T. muenninki cDNA clones of the Y-linked genes.
Materials and methods
Sample collection, cell culture, and DNA and RNA extraction
The three species of Tokudaia are all endangered (The IUCN Red List of Threatened Species; http://www.iucnredlist.org/ 15/9/2011) and have been protected by the Japanese government as natural treasures since 1972. With permission from the Agency for Cultural Affairs and the Ministry of the Environment in Japan, we trapped the Okinawa spiny rat, T. muenninki, on Okinawa-jima Island in March 2008 and February 2009 (Yamada et al. 2010). We collected tail tissue from four males and three females for use in the study. Cell culture and DNA extraction were performed as described by Murata et al. (2010). Total RNA was extracted from fibroblasts derived from tail tissue from a T. muenninki male using TRIZOL Reagent (Invitrogen) in accordance with the manufacturer’s protocol. The cDNAs were synthesized using SuperScript II Reverse Transcriptase (Invitrogen).
DNA probes
Seven mouse cDNA clones of functional genes linked to chromosome 11 and 22 linked to the X chromosome were used in the study. Their gene names, symbols, accession numbers, sizes, and chromosomal location in mouse (M. musculus), rat (Rattus norvegicus), and three Tokudaia species are listed in Table 1. A 6.6-kilo base pairs (kb) mouse genomic DNA fragment was used for chromosomal localization of the 18S–28S rRNA gene (Kominami et al. 1982).
Clones of T. muenninki DDX3Y (AB672502; 1.8 kb) and UTY (AB672501; 2.0 kb), which are both linked to the Y chromosome, were also used for FISH mapping. The mouse homologues referred to here are Ddx3y (NM_012008) and Uty (NM_009484), and the rat genes are DDX3Y (FJ775727) and UTY (FJ775728). The T. muenninki homologues of these genes were cloned by degenerate PCR. The primer pairs used in the PCR were as follows: DDX3Y, 5′-TGA TCG TGG AAG TGG ATC C-3′and 5′-CAC AGC TGG TAA TGT ACT GCA-3′; UTY, 5′-GAC GCT GTT GAA CAA GGC AA-3′ and 5′-GGA GTG GGT GGA TTC AAC TTC-3′. The PCR for DDX3Y was carried out in 10 μl of 1× Ex Taq buffer that contained 0.5 μl of template cDNA, 2 mM MgCl2, 0.5 μM of each primer, 0.2 mM each of the four dNTPs, and 0.25 U of Ex Taq (TaKaRa Bio Inc.). The reaction profile was denaturation for 5 min at 94°C, followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 60 °C, and extension for 2 min at 72°C, and then a final extension at 72°C for 7 min. The PCR for UTY was carried out in 20 μl of 1× PrimeSTAR GXL buffer (Mg2+ plus) that contained 1.0 μl of template cDNA, 0.3 μM of each primer, 0.2 mM each of the four dNTPs, and 0.25 U of PrimeSTAR GXL DNA Polymerase (TaKaRa Bio Inc.). The reaction profile was denaturation for 3 min at 94°C, followed by 35 cycles of denaturation for 10 s at 98°C, annealing for 15 sec at 60°C, and extension for 1.5 min at 68°C, and then a final extension at 72°C for 5 min.
Chromosome preparation and Zoo-FISH analysis
The preparation of R-banded chromosomes was performed as described by Matsuda et al. (1992a), Matsuda and Chapman (1995), and Kobayashi et al. (2008).
The Zoo-FISH analysis was performed using biotin- and Cy3-labeled chromosome-specific painting probes from the laboratory mouse (Cambio Ltd.). FISH was carried out as described by Nakamura et al. (2007) with slight modifications. The probes were denatured at 75°C for 10 min and pre-annealed by incubation at 37°C for 1 h. The chromosome slides were hybridized with the biotin- or Cy3-labeled probes at 37°C for 5 days. After hybridization with the biotin-labeled probes, the slides were washed in 50% formamide in 2× saline sodium citrate (SSC) at 37°C for 15 min, in 2× SSC for 15 min, and then in 4× SSC for 5 min at room temperature. To detect the hybridization signals of the biotin-labeled probes, the chromosome slides were incubated with avidin fluorescein isothiocyanate (FITC) (Vector Laboratories) or streptavidin-Cy5 (GE Healthcare UK Ltd.) diluted 1:500 in 1% BSA/4× SSC at 37°C for 1 h. The slides were washed sequentially on a shaker with 4× SSC, 0.1% Nonidet P-40/4× SSC, and then 4× SSC for 5 min each at room temperature. After hybridization with the Cy3-labeled probe, the slides were washed sequentially in 4× SSC, 0.1% Nonidet P-40/4× SSC, and then 4× SSC for 5 min each at room temperature.
CGH analysis
The CGH analysis was conducted using metaphase spreads from one male and one female of T. muenninki as described by Kobayashi et al. (2007) with slight modifications. The genomic DNA for the probes was also extracted from one male and one female. The male and female genomic DNA probes were labeled differently, with Cy3-deoxyuridine 5-triphosphate (dUTP; GE Healthcare) and fluorescein-12-dUTP (Invitrogen), respectively, by nick translation at 15°C for 90 min. The labeled DNA probes were denatured at 75°C in formamide and dissolved in hybridization buffer (50% formamide, 2× SSC, 10% dextran sulfate, and 2 mg/ml BSA). The cocktail of probes for one slide contained 700 ng of labeled DNA from the female and 700 ng of labeled DNA from the male. Slides were hybridized at 37°C for 4 days. The slides were then washed sequentially in 4× SSC, 0.1% Nonidet P-40 in 4× SSC, 4× SSC, and 2× SSC for 5 min each at room temperature, and then rinsed in 2× SSC.
FISH analysis
The FISH analysis using cDNA clones as probes was performed as described by Kuroiwa et al. (2010). The exception was the FISH analysis using the 18S–28S rRNA gene clone as a probe. In this case, the chromosome slides were treated with RNase (1 μg/μl in 2× SSC) for 1 h, washed three times in 2× SSC for 3 min each, and dehydrated in 70% and 100% ethanol at 4°C for 5 min each before the chromosomes were denatured in 70% formamide in 2 × SSC at 69°C for 2 min. After hybridization and washing, the slides were incubated under parafilm for 1 h at 37°C with fluoresceinated avidin (avidin-FITC; Vector Laboratories) diluted 1:500 in 1% bovine serum albumin (BSA)/4× SSC [instead of goat anti-biotin antibody (Vector Laboratories)]. The slides were washed sequentially on a shaker with 4× SSC, 0.1% Nonidet P-40 in 4× SSC, and 4× SSC for 10 min each, and then rinsed with 2× SSC and stained with 0.75 mg/ml propidium iodide for 3 min.
Microphotography and image capture
The chromosomal slides were observed under a Nikon fluorescence microscope using Nikon filter sets B-2A (450–490 nm) and ultraviolet (UV)-2A (330–380 nm). Kodak Ektachrome American Standards Association (ASA) 100 films were used for microphotography. The digital FISH images were captured using the 550CW-QFISH application program of Leica Microsystems Imaging Solution Ltd. using a cooled CCD camera (MicroMAX 782Y, Princeton Instruments) mounted on a Leica DMRA microscope.
Results
Zoo-FISH analysis
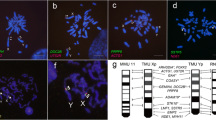
We conducted the Zoo-FISH analysis on chromosomes from a male and a female specimen of T. muenninki using all M. musculus chromosome (MMU)-specific painting probes except for the Y chromosome probe. Each chromosome of T. muenninki could be identified on the basis of the Hoechst-stained bands that were obtained by the replication R-banding method (Fig. 1). The MMU probes for all autosomes and the X chromosome were hybridized successfully to T. muenninki chromosomes (TMU). The images for the probes MMUX, MMU11, and MMU16, are shown in Fig. 1a–c. The patterns for all the T. muenninki chromosomes are summarized in Fig. 1f. Although the long arm of the X chromosome of T. muenninki was covered with the MMUX probe (Fig. 1a), the short arms of the X and Y chromosomes both hybridized with the mouse probes MMU11 and MMU16 (Fig. 1b and c, respectively). The 18S–28S ribosomal RNA genes were localized to the distal ends of chromosome 1 and the short arms of the sex chromosomes (Fig. 1d, e), which corresponded to MMU9 and MMU11, respectively (Fig. 1f).
Karyotypic characterization in T. muenninki. a–c Zoo-FISH for T. muenninki with MMU-specific probes. Hybridization of FITC- (green) labeled mouse painting probes, MMUX, to PI-stained chromosomes of a T. muenninki female (a). Hybridization of Cy3- (yellow) and Cy5- (pink) labeled mouse painting probes, MMU11 (b) and MMU 16 (c), respectively, to Hoechst-stained chromosomes of a T. muenninki male. Arrows and arrowheads indicate the hybridization signals on the sex chromosomes and autosomes [TMU10 (b) and TMU15 (c)], respectively. The scale bar indicates 10 μm. (d, e) Mapping of T. muenninki chromosomes by FISH using mouse genomic DNA clones of the 18S–28S rRNA genes. Hybridization signals of the 18S–28S rRNA genes were visualized by avidin-FITC on PI-stained chromosomes (d). Hoechst G-banding patterns are shown in e. Arrows and arrowheads indicate the hybridization signals on the sex chromosomes and a pair of autosomes (TMU1), respectively. The scale bar indicates 10 μm. f Comparative cytogenetic maps of T. muenninki. The comparative cytogenetic maps showing chromosome homologies between mouse and T. muenninki were constructed by Zoo-FISH analysis with mouse probes. The numbers under the chromosomes of T. muenninki indicate the chromosome numbers for this species. The numbers inside the chromosomes indicate the chromosome numbers for mouse that correspond to the chromosomal segments of T. muenninki indicated. Arrowheads indicate the locations of the 18S–28S rRNA genes. Large heterochromatic regions are shown in black
Fourteen chromosomes of T. muenninki (TMU1, 2, 3, 5, 7, 9, 11, 14, 15, 17, 18, 19, 20, and 21) each were hybridized with a single mouse probe; six chromosomes (TMU4, 6, 8, 10, 12, and 13) each hybridized with two probes; and one chromosome (TMU16) was hybridized with four probes (Fig. 1f). Thirty-four conserved segments were detected between the mouse and T. muenninki chromosomes. The hybridization patterns between T. muenninki and the mouse probes could be grouped into three categories: (1) 11 mouse probes (MMU2, 3, 4, 6, 7, 9, 12, 14, 18, 19, and X) each hybridized to a single chromosome or chromosomal segment of T. muenniki; (2) six mouse probes (MMU1, 8, 11, 13, 15, and 16) each hybridized to two chromosomes; and (3) three mouse probes (MMU5, 10, and 17) each hybridized to more than three chromosomes (Fig. 1f).
CGH analysis
We performed CGH to identify the male-specific region in the Y chromosome of T. muenninki. CGH signals with differently labeled genomic DNAs from a female and a male were highlighted in the long arm of the Y chromosome, as well as in the heterochromatic regions of five autosomal pairs in both sexes (Fig. 2). The long arm of the Y chromosome, Yq, was labeled intensely not only by the male probe, which contained Y chromosomal DNA, but also by the female probe, although the intensity of Hoechst fluorescence of Yq was not higher than that of any of the other chromosomes (Fig. 2a–c). The short arm of the Y chromosome, Yp, which is a proto-autosomal segment that has been translocated to the Y chromosome, as shown in Fig. 1, was stained equally by the female- and male-derived probes (Fig. 2d). Therefore, the loss or gain of a male-specific chromosomal region was not detected in the T. muenninki male by CGH analysis. Furthermore, when unlabeled female DNA and mouse C0t-1 DNA were used in the CGH analysis as competitors, no male-specific region was detected in the T. muenninki male (data not shown). In addition, the sex-specific region was not detected in diploid mitotic complements of the T. muenninki female (Fig. 2e–h).
CGH images of a male (a–d) and a female (e–h) metaphase spread for T. muenninki. The Hoechst G-banding patterns are shown in a and e. The CGH images (d, h) were obtained by merging the images of metaphase spreads hybridized with female genomic DNA (b, f) and male genomic DNA (c, g). Arrows indicate the sex chromosomes. Scale bar represents 10 μm
Comparative FISH and FISH analysis
We mapped mouse cDNA clones of 22 X-linked genes on chromosomes derived from a T. muenniki female by direct R-banding FISH. All 22 genes were mapped on the long arm of the X chromosome in T. muenninki. The gene order was identical to that of the other two Tokudaia species and the laboratory rat, R. norvegicus (Fig. 3, Kobayashi et al. 2008). The hybridization patterns of four genes on the X chromosome, ELK1 (Fig. 4a, b), SAT1 (Fig. 4c, d), SLC25A5 (Fig. 4e, f), and PLXNA3 (Fig. 4g, h), are shown in Fig. 4.
Comparative cytogenetic maps of seven genes from mouse chromosome 11 on the sex chromosomes of T. muenninki (TMUX, TMUY) and mouse chromosome 11 (MMU11) and 22 X-linked genes in T. muenninki and T. tokunoshimensis. The locations and order of the genes are shown on the side of each chromosome. The gene order and ideogram of MMU11 was taken from the NCBI database (http://www.ncbi.nlm.nih.gov/ 21/9/2011). The gene order and ideograms of T. tokunoshimensis X chromosomes were taken from Kobayashi et al. (2008). TTO, T. tokunoshimensis chromosome
Chromosome mapping of four X-linked genes and two Y-linked genes on T. muenninki chromosomes. The hybridization signals are indicated by arrows. Four X-linked genes, ELK1 (a, b), SAT1 (b, c), SLC25A5 (e, f), and PLXNA3 (g, h), were mapped from the central to the distal part of T. muenninki Xq in the order shown. Two Y-linked genes, DDX3Y (i, j) and UTY (k, l) were localized to the proximal region of the centromere in T. muenninki Yp. R- and Hoechst G-banding patterns are shown in (a, c, e, g, i, k) and (b, d, f, h, j, l), respectively. The scale bar indicates 10 μm
Mapping analysis of the SRY gene could not be used to identify the original Y chromosome region because multiple copies of SRY are distributed over the entire Yq and centromere region of the Y chromosome in T. muenninki (Murata et al. 2010). In light of this, we used nine mouse cDNA clones (unpublished data) to screen for single copy genes on the Y chromosome by Southern blot analysis. The DDX3Y and UTY genes appeared to occur as a single copy in the male T. muenninki genome. Thus, we cloned the T. muenninki cDNA for both DDX3Y and UTY and used them as probes for FISH. The sequence identity of the DDX3Y and UTY genes between mouse and T. muenninki was 95%. Both the DDX3Y and UTY genes were localized in T. muenninki to the proximal region of the centromere on Yp (Fig. 4i–l).
Given that the MMU11 painting probe hybridized to both Xp and Yp of T. muenninki, we performed comparative FISH mapping using probes from mouse cDNA clones of seven genes located on chromosome 11. All seven genes mapped to both Xp and Yp in T. muenninki. The hybridization patterns of four genes, STK10 (Fig. 5a–d), DOC2B (Fig. 5e–h), COASY (Fig. 5i–l), and CBX2 (Fig. 5m–o), are shown in Fig. 5. The gene orders on both Xp and Yp were identical to that on chromosome 11 of mouse (Fig. 3). However, CBX2 was also localized to chromosome 14 in both sexes of T. muenniki (Fig. 5m–o).
Chromosome mapping of the STK10 (a–d), DOC2B (e–h), COASY (i–l), and CBX2 (m–o) genes on T. muenninki chromosomes. The STK10, DOC2B, and COASY genes were localized to both Xp (a, e, i) and Yp (c, g, k) in T. muenninki. CBX2 was mapped to Xp, Yp, and a pair of autosomes (TMU14) in the male (m) and Xp and TMU14 in the female (o). The hybridization signals are indicated by arrows on the sex chromosomes and arrowheads on the autosomes. R- and Hoechst G-banding patterns are shown in (a, c, e, g, i, k, m, o) and (b, d, f, h, j, l, n), respectively. The scale bar represents 10 μm
Discussion
Ancestral karyotype of the genus Tokudaia
With the exception of several chromosome rearrangements described below, the karyotype of T. muenninki was largely identical to the one reported previously, which was predicted from maps of T. osimensis and T. tokunoshimensis generated by chromosome painting (Nakamura et al. 2007): MMU1b/17a/17e, a single segment of MMU14, and the two distinct chromosomal segments of MMU15 occurred in T. muenninki, whereas MMU1b/17a, two distinct chromosomal segments of MMU14, and a single segment of MMU15 occurred in T. osimensis and T. tokunoshimensis. The former segments found in T. muenninki are also conserved in the ancestral karyotype of the genus Apodemus (Matsubara et al. 2004), which is the most closely related group to the genus Tokudaia in Murinae (Michaux et al. 2002; Sato and Suzuki 2004; Rowe et al. 2008), suggesting that the ancestor of genus Tokudaia might have the same segments as T. muenninki, MMU1b/17a/17e, two distinct chromosomal segments of MMU14 and a single segment of MMU15. Therefore, we have demonstrated that the ancestral karyotype of the genus Tokudaia had a diploid chromosome number of 48 and contained the following chromosomes that are homologous to mouse: 1a, 1b/17a/17e, 2, 3, 4, 5a, 5b/11a, 5c/6, 7/19, 8a, 8b, 9, 10a, 10b/17b, 10c/17c, 11b/16a, 12/17d, 13a, 13b/15a, 14, 15b, 16b, 18, X, and Y (Fig. 6). Lineage-specific chromosome rearrangements in T. muenninki, inferred from the ancestral karyotype, were as follows: (1) fusion between segments homologous to MMU11b/16a and the ancestral Y in TMUY; (2) centric fusions between segments homologous to MMU11b/16a and MMUX in TMUX and between segments homologous to MMU10b/17b and MMU13b/15a in TMU16; and (3) two pericentric inversions in TMU17 and TMU19 (Fig. 6).
A schematic representation of the ancestral karyotype of the Tokudaia species and chromosome rearrangements that occurred in T. muenninki after its divergence from the common ancestor. The numbers inside the chromosomes indicate the mouse chromosome numbers that correspond to the indicated chromosomal segments of the Tokudaia species. The lower chromosomes correspond to the chromosomes of T. muenninki, and the numbers underneath are the chromosome numbers for this species. The numbers above the upper chromosomes, those of the schematic ancestral karyotype, indicate the chromosomes, chromosomal segments, or chromosome associations that are homologous to mouse chromosomes. They were numbered following the nomenclature of Stanyon et al. (2004) and Nakamura et al. (2007). Arrows show how the chromosomes of T. muenninki were derived from the ancestral karyotype. Two-pronged arrows to a single chromosome indicate the successive occurrence of fusions. “Inv” indicates a pericentric inversion
Furthermore, the localization of the 18S–28S ribosomal RNA genes to two chromosomal segments homologous to MMU9 and MMU11 in T. muenninki (Fig. 1d–f) was also conserved in the other two Tokudaia species (Arakawa et al. 2002; Nakamura et al. 2007) and six Apodemus species (Matsubara et al. 2004), which suggested that no chromosomal variation had occurred in T. muenninki in these regions.
Fusion of autosomes and the sex chromosomes
The short arms of the X and Y chromosomes of T. muenninki were painted by the MMU11 and MMU16 probes, which indicated that fusions had occurred between a pair of autosomes and the sex chromosomes. This was not observed in the other two Tokudaia species, where the chromosomal segments recognized by MMU11 and MMU16 corresponded to chromosome 3q in T. osimensis and chromosome 10 in T. tokunoshimensis, respectively (Nakamura et al. 2007). The fusion of a pair of autosomes with both the X and Y chromosomes is a rare chromosomal rearrangement in mammals. The only other reported cases are in: the giant mole-rat (Fukomys mechowii), the African pygmy mouse (Mus minutoides), and three bovine species (Boselaphus tragocamelus, Tragelaphus spekei, and Tragelaphus imberbis) (Deuve et al. 2006; Veyrunes et al. 2004, 2010; Gallagher et al. 1998; Rubes et al. 2008). Species with X-autosome translocations all have intercalary heterochromatic blocks (IHBs) between the autosomal and the ancestral sexual segments, which probably enables the timing of replication of the sex and autosomal segments to be regulated independently (Dobigny et al. 2004). The centromeric heterochromatin block of T. muenninki might function as an IHB and an X-autosomal boundary in this species (Murata et al. 2010).
Avoidance of Y chromosome loss through fusion with an autosome
T. muenninki is known to be the only species with a Y chromosome in the genus Tokudaia; as a consequence, it follows the general XY pattern of mammalian sex determination (Tsuchiya et al. 1989). However, our results indicated that the Y chromosome of T. muenninki has evolved in a unique manner. The Y chromosome might have had instability persisting in the common ancestral species of the genus Tokudaia (Fig. 7). In an ancestral lineage common to the two XO/XO spiny rats, T. osimensis and T. tokunoshimensis, most of the Y-linked genes (except SRY and RBMY1A1) escaped to the X chromosome, and the Y chromosome was lost subsequently (Kuroiwa et al. 2010). In contrast, in the ancestral population of T. muenninki, the X and Y chromosomes fused with a pair of autosomes.
The evolution of sex chromosomes, SRY, and CBX2 in the genus Tokudaia. a The evolutionary events inferred from the present study (red) and previous studies (black) are shown in the phylogeny, together with the geographical distribution of Tokudaia species. T. muenninki, T. osimensis, and T. tokunoshimensis inhabit only Okinawa-jima, Amami-Osima, and Tokunoshima Islands, respectively. b, c Adult female (b) and juvenile to sub-adult male (c) of T. muenninki trapped as part of conservation activities
The enlargement of the Y chromosome through fusion with an autosome extended the pseudoautosomal regions (PARs). PARs behave like an autosomal pair and recombine during male meiosis, which is thought to play a critical role in spermatogenesis (Kauppi et al. 2011; Matsuda et al. 1992b; Mohandas et al. 1992). The XY male mice with inverted X PAR, Y PAR flanked at the distal end by X PAR boundary together with adjacent X-specific material, or both of the variant PARs, are known to produce XO progeny by leading to the production of unusual X and Y products and consequent sex chromosome loss (Burgoyne and Evans 2000). If variant PAR was associated with the Y chromosome loss in an ancestral lineage common to the two XO/XO spiny rats, T. osimensis and T. tokunoshimensis, the Y chromosome would be stabilized by the acquisition of new PARs in the ancestral lineage of T. muenninki, and thus, loss of the Y chromosome was avoided. To test this hypothesis, future research should include the identification and sequencing of an ancestral PAR in three Tokudaia species.
Most human Y-linked genes are relics of a recent autosomal addition (Waters et al. 2001). The PAR of placental mammals was part of a larger autosomal addition to the X and Y chromosomes 130–80 million years ago (MYA) (Toder and Graves 1998), which suggests that the ancestral PAR of placental mammals was larger than the present human PAR (Waters et al. 2007). Enlargement of the Y chromosome by the addition of an autosome could be an important chromosomal phenomenon to stabilize X–Y pairing in the lineage of placental mammals.
Structure of the giant X and Y chromosomes
All mouse cDNA clones of the X-linked genes were mapped to Xq (Fig. 3). The gene order was consistent with that of rat and the two other Tokudaia species (Kobayashi et al 2008), which suggests that the proto-X chromosome was retained in Xq and had not undergone chromosomal rearrangement, e.g., inversion and deletion on a broad scale.
In our CGH analysis, probes derived from either males or females hybridized to Yq of T. muenninki (Fig. 2). This might have been caused by the accumulation of repetitive sequences on Yq that were common to both sexes. The accumulation of numerous repetitive sequences could be the reason why a male-specific region was not detected clearly by CGH, even though many SRY sequences are known to occur on Yq of T. muenninki (Murata et al. 2010).
Previous attempts to map the SRY gene did not detect an ancestral Y segment that was common to other mammalian Y chromosomes due to the large number of mainly nonfunctional SRY copies. In the present study, using FISH to map two single-copy Y-linked genes, DDX3Y and UTY, we identified the region of Yp close to the centromere as the euchromatic region of the proto-Y chromosome in the genus Tokudaia (Fig. 4). Now that this region has been identified, further characterization of the Y-linked genes in T. muenninki by Southern blotting and FISH analyses is needed to reveal the gene content of the original Y region. Sequence determination of the region using a bacterial artificial chromosome library is also required to determine the gene order and gene structure of the putative ancestral Y chromosome in T. muenninki.
Neo-X? Neo-Y?
Comparative FISH mapping of seven mouse cDNA clones linked to chromosome 11 showed that the order of genes on T. muenninki Xp and Yp was identical to that of mouse chromosome 11. No obvious chromosome rearrangement, e.g., inversion, was detected between the X and Y chromosomes. However, it remains possible that some divergence in nucleotide sequence has occurred between the X and Y chromosomes if the proto-autosomal segments are in the early stages of differentiation. On the basis of mitochondrial sequence data for cytochrome b, divergence times between T. muenninki and the two other species are estimated to be around 2.5–2.7 MYA (Murata et al. 2010; on the basis of the substitution rate of this gene in murids, 4.8% per MY; Suzuki et al. 2003). In the black muntjac (Muntiacus crinifrons), some sequence divergence has occurred between neo-sex chromosomes within the past 0.5 MY (Zhou et al. 2008). Therefore, there has been sufficient time for mutations to accumulate between the sex chromosomes of T. muenninki.
Duplication of CBX2 in the genus Tokudaia
Gene duplication has been proposed as a primary source of material for the acquisition of evolutionary novelties, e.g., new gene functions and new patterns of expression (Ohno 1970; Sidow 1996). Several mechanisms have been put forward to explain how paralogs diverge from one another. In one process, asymmetric evolution, one gene copy retains the ancestral function through purifying selection, whereas the other copy can undergo neofunctionalization or pseudogenization (Lynch and Conery 2000). Such a duplication event allowed the evolution of a sex-determining gene in a model fish species, medaka (Oryzas latipes, Schartl 2004).
Previously, we examined the copy numbers and chromosomal locations of ten genes, including CBX2, to identify candidate genes for sex determination in the two Tokudaia species that lack SRY (Kuroiwa et al. 2011). The Cbx2/CBX2 gene is a single copy gene in mice and humans and is known be involved in gonadal differentiation. The knockout of Cbx2 in mouse causes male-to-female sex reversal, and that mouse with the karyotype 40, XY has normal female external genitalia, which indicates that Cbx2/CBX2 represses ovarian development (Katoh-Fukui et al. 1998; Biason-Lauber et al. 2009). CBX2 occurs at two loci in T. osimensis and T. tokunoshimensis, owing to the translocation of a duplicated copy of the gene to another autosome. Furthermore, two or three more copies of CBX2 are present in males of these species as compared with females. It is likely that these extra copies of CBX2 repress ovarian development and hence cause the undifferentiated gonad to develop testes in these species. Therefore, the differences in gene dosage between sexes might be involved in a novel sex-determining mechanism (Kuroiwa et al. 2011). In the present study, we have shown that T. muenninki also possesses multiple copies of CBX2. This suggests that the duplication of CBX2 occurred in a common ancestor of all three Tokudaia species, after which the autosomal segment that contained the original CBX2 copy became linked to the T. muenninki sex chromosomes through chromosomal fusion. The comparison of the copy number of CBX2 between T. muenninki male and female is needed to reveal when Tokudaia species acquired the extra copies of CBX2 in male genome.
T. muenninki possesses multiple copies of the SRY gene on the Y chromosome (Murata et al. 2010). Most of the copies are pseudogenes, with a complete ORF conserved in only three copies (Murata et al. 2010). A mutation, which results in a single amino acid substitution in the DNA binding surface domain of the HMG box of all SRY copies, might have weakened the DNA binding ability of the SRY protein in T. muenninki. If the Y-linked CBX2 in T. muenninki has acquired a male-specific function through early differentiation between Xp and Yp, CBX2 might also have become an important gene for sex determination in T. muenninki in the presence of a weakened SRY. Future research should include the sequencing of each copy of CBX2 and a functional analysis of the SRY genes in T. muenninki to test this hypothesis.
Abbreviations
- ASA:
-
American Standards Association
- BSA:
-
Bovine serum albumin
- cDNA:
-
Complementary DNA
- CGH:
-
Comparative genomic hybridization
- dUTP:
-
Deoxyuridine 5-triphosphate
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- HMG:
-
High-mobility group
- IHB:
-
Intercalary heterochromatin block
- kb:
-
Kilo base pairs
- MMU:
-
Mus musculus chromosome
- MYA:
-
Million years ago
- ORF:
-
Open reading frame
- PAR:
-
Pseudoautosomal region
- qRT-PCR:
-
Quantitative real-time PCR
- RBMY1A1 :
-
RNA binding motif protein, Y chromosome, family 1, member A1
- SSC:
-
Saline sodium citrate
- TMU:
-
Tokudaia muenninki chromosome
- TTO:
-
Tokudaia tokunoshimensis chromosome
- UV:
-
Ultraviolet
- Zoo-FISH:
-
Cross-species fluorescence in situ hybridization
References
Arakawa Y, Nishida-Umehara C, Matsuda Y, Sutou S, Suzuki H (2002) X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet Genome Res 99:303–309
Biason-Lauber A, Konrad D, Meyer M, Beaufort C, Schoenle EJ (2009) Ovaries and female phenotype in a girl with 46, XY karyotype and mutations in the CBX2 gene. Am J Hum Genet 84:658–663
Burgoyne PS, Evans EP (2000) A high frequency of XO offspring from XPaf Y* male mice: evidence that the Paf mutation involves an inversion spanning the X PAR boundary. Cytogenet Cell Genet 91:57–61
Deuve JL, Bennett NC, O’Brien PCM et al (2006) Complex evolution of X and Y autosomal translocations in the giant mole-rat, Cryptomys mechowi (Bathyergidae). Chromosome Res 14:681–691
Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V (2004) Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study. Chromosoma 113:34–41
Elliott DJ (2004) The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl 27:328–334
Gallagher DS, Davis SK, De Donato M et al (1998) A karyotypic analysis of nilgai, Boselaphus tragocamelus (Artiodactyla: Bovidae). Chromosome Res 6:505–513
Honda T, Suzuki H, Itoh M (1977) An unusual sex chromosome constitution found in the Amami spinous country-rat, Tokudaia osimensis osimensis. Jpn J Genet 52:247–249
Honda T, Suzuki H, Itoh M, Hayashi K (1978) Karyotypical differences of the Amami spinous country-rats, Tokudaia osimensis osimensis obtained from two neighbouring islands. Jpn J Genet 53:297–299
Katoh-Fukui Y, Tsuchiya R, Shiroishi T et al (1998) Male-to-female sex reversal in M33 mutant mice. Nature 393:688–692
Kauppi L, Barchi M, Baudat F et al (2011) Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331:916–920
Kobayashi T, Yamada F, Hashimoto T et al (2007) Exceptional minute sex-specific region in the X0 mammal, Ryukyu spiny rat. Chromosome Res 15:175–187
Kobayashi T, Yamada F, Hashimoto T et al (2008) Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res 16:587–593
Kominami R, Mishima Y, Urano Y, Sakai M, Muramatsu M (1982) Cloning and determination of the transcription termination site of ribosomal RNA gene of the mouse. Nucl Acids Res 10:1963–1979
Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351:117–121
Kuramochi S, Matsuda Y, Okamoto M et al (1999) Molecular cloning of the human gene STK10 encoding lymphocyte-oriented kinase, and comparative chromosomal mapping of the human, mouse, and rat homologues. Biochem Biophys Res Commun 270:522–527
Kurohara K, Matsuda Y, Nagabukuro A, Tsuji A, Amagasa T, Fujisawa-Sehara A (2000) Meltrin beta (ADAM19) gene: cloning, mapping, and analysis of the regulatory region. Immunogenetics 49:369–375
Kuroiwa A, Watanabe T, Hishigaki H, Takahashi E, Namikawa T, Matsuda Y (1998) Comparative FISH mapping of mouse and rat homologues of twenty-five human X-linked genes. Cytogenet Cell Genet 1:208–212
Kuroiwa A, Ishiguchi Y, Yamada F, Abe S, Matsuda Y (2010) The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma 119:519–526
Kuroiwa A, Handa S, Nishiyama C et al (2011) Additional copies of CBX2 in the genomes of males of mammals lacking SRY, the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis). Chromosome Res 19:635–644
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y (2004) Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res 12:383–395
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matsuda Y, Harada Y-N, Natsuume-Sakai S et al (1992a) Location of the mouse complement factor H gene (cfh) by FISH analysis and replication R-banding. Cytogenet Cell Genet 61:282–285
Matsuda Y, Moens PB, Chapman VM (1992b) Deficiency of X and Y chromosomal pairing at meiotic prophase in spermatocytes of sterile interspecific hybrids between laboratory mice (Mus domesticus) and Mus spretus. Chromosoma 101:483–492
Mazeyrat S, Saut N, Mattei MG, Mitchell MJ (1999) RBMY evolved on the Y chromosome from a ubiquitously transcribed X-Y identical gene. Nat Genet 22:224–226
Michaux JR, Chevret P, Filippucci MG, Macholan M (2002) Phylogeny of the genus Apodemus with a special emphasis on the subgenus Sylvaemus using the nuclear IRBP gene and two mitochondrial markers: cytochrome b and 12S rRNA. Mol Phylogenet Evol 23:123–136
Mohandas TK, Speed RM, Passage MB et al (1992) Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: meiotic studies in a man with a deletion of distal Xp. Am J Hum Genet 51:526–533
Murata C, Yamada F, Kawauchi N, Matsuda Y, Kuroiwa A (2010) Multiple copies of SRY on the large Y chromosome of the Okinawa spiny rat, Tokudaia muenninki. Chromosome Res 18:623–634
Nakamura T, Kuroiwa A, Nishida-Umehara C, Matsubara K, Yamada F, Matsuda Y (2007) Comparative chromosome painting map between two Ryukyu spiny rat species, Tokudaia osimensis and Tokudaia tokunoshimensis (Muridae, Rodentia). Chromosome Res 15:799–806
Ohno S (1970) Evolution by gene duplication. Springer, Berlin–Heidelberg–New York
Rowe KC, Reno ML, Richmond DM, Adkins RM, Steppan SJ (2008) Pliocene colonization and adaptive radiations in Australia and New Guinea (Sahul): Multilocus systematics of the old endemic rodents (Muroidea: Murinae). Mol Phylogenet Evol 47:84–101
Rubes J, Kubickova S, Pagacova E et al (2008) Phylogenomic study of spiral-horned antelope by cross-species chromosome painting. Chromosome Res 16:935–947
Sato JJ, Suzuki H (2004) Phylogenetic relationships and divergence times of the genus Tokudaia within Murinae (Muridae; Rodentia) inferred from the nucleotide sequences encoding the Cytb gene, RAG1, and IRBP. Can J Zool 82:1343–1351
Schartl M (2004) Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev 14:634–641
Sidow A (1996) Gen(om ) e duplications in the evolution of early vertebrates. Curr Opin Genet Dev 6:715–722
Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346:240–244
Soullier S, Hanni C, Catzeflis F, Berta P, Laudet V (1998) Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm Genome 9:590–592
Stanyon R, Yang F, Morescalchi AM, Galleni L (2004) Chromosome painting in the long-tailed field mouse provides insights into the ancestral murid karyotype. Cytogenet Genome Res 105:406–411
Sutou S, Mitsui Y, Tsuchiya K (2001) Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm Genome 12:17–21
Suzuki H, Sato JJ, Tsuchiya K et al (2003) Molecular phylogeny of wood mice (Apodemus, Muridae) in East Asia. Biol J Linn Soc 80:469–481
Toder R, Graves JA (1998) CSF2RA, ANT3, and STS are autosomal in marsupials: implications for the origin of the pseudoautosomal region of mammalian sex chromosomes. Mamm Genome 9:373–376
Tsuchiya K, Wakana S, Suzuki H, Hattori S, Hayashi Y (1989) Taxonomic study of Tokudaia (Rodentia: Muridae): I. Genetic differentiation. Memoirs Nat Sci Museum. Tokyo 22:227–234
Veyrunes F, Catalan J, Sicard B et al (2004) Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Murinae; Mus). Chromosome Res 12:369–382
Veyrunes F, Catalan J, Tatard C et al (2010) Mitochondrial and chromosomal insights into karyotypic evolution of the pygmy mouse, Mus minutoides, in South Africa. Chromosome Res 18:563–574
Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JA (2001) The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet 92:74–79
Waters PD, Wallis MC, Marshall Graves JA (2007) Mammalian sex—origin and evolution of the Y chromosome and SRY. Semin Cell Dev Biol 18:389–400
Yamada F, Kawauchi N, Nakata K et al (2010) Rediscovery after thirty years since the last capture of the critically endangered Okinawa spiny rat Tokudaia muenninki in the northern part of Okinawa Island. Mammal Study 35:243–255
Zhou Q, Wang J, Huang L et al (2008) Neo-sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol 9:R98
Acknowledgments
The authors thank C. Nishida-Umehara, K. Matsubara, M. Tsuji. K. Sahara, and A. Yoshido for help with cell culture and FISH. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a grant from the Naito Foundation, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Tariq Ezaz and Jennifer Graves.
Rights and permissions
About this article
Cite this article
Murata, C., Yamada, F., Kawauchi, N. et al. The Y chromosome of the Okinawa spiny rat, Tokudaia muenninki, was rescued through fusion with an autosome. Chromosome Res 20, 111–125 (2012). https://doi.org/10.1007/s10577-011-9268-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9268-6