Abstract
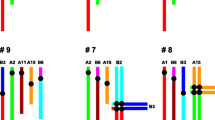
X-autosome translocations are highly deleterious chromosomal rearrangements due to meiotic disruption, the effects of X-inactivation on the autosome, and the necessity of maintaining different replication timing patterns between the two segments. In spite of this, X-autosome translocations are not uncommon. We here focus on the genus Taterillus (Rodentia, Gerbillinae) which provides two sister lineages differing by two autosome–gonosome translocations. Despite the recent and dramatic chromosomal repatterning characterising these lineages, the X-autosome translocated species all display intercalary heterochromatic blocks (IHBs) between the autosomal and the ancestral sexual segments. These blocks, composed of highly amplified telomeric repeats and rDNA clusters, are not observed on the chromosomes of the non-translocated species, nor the Y1 and Y2 of the translocated species. Such IHBs are found in all mammals documented for X-autosome translocation. We propose an epigenomic hypothesis which explains the viability of X-autosome translocations in mammals. This posits that constitutive heterochromatin is probably selected for in X-autosome translocations since it may (1) prevent facultative heterochromatinization of the inactivated X from spreading to the autosomal part, and (2) allow for the independent regulation of replication timing of the sex and autosomal segments.




Similar content being viewed by others
References
Aagaard L, Schmid M, Warburton P, Jenuwein T (2000) Mitotic phosphorylation of Suv39H1, a novel component of active centromere, coincides with transient accumulation at mammalian centromeres. J Cell Sci 113:817–829
Ashley T (2002) X-autosome translocations, meiotic synapsis, chromosome evolution and speciation. Cytogenet Genome Res 96:33–39
Avner D, Heard E (2001) X-chromosome inactivation: counting, choice and initiation. Nat Genet 2:59–67
Bailey JA, Carrel L, Chakravarti A, Eichler EE (2000) Molecular evidence for a relationship between LINE-1 elements and chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA 97:6634–6639
Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo-domain. Nature 410:120–124
Bell AC, West AG, Felsenfeld G (2001) Insulators and boundaries: versatile elements in the eukaryotic genome. Science 291:447–450
Benazzou T (1984) Contribution à l’étude chromosomique et de la diversification biochimique des Gerbillidés (Rongeurs). PhD thesis. Université de Paris XI, Orsay
Boggs BA, Chinault AC (1994) Analysis of replication timing properties of human X-chromosomal loci by fluorescence in situ hybridization. Proc Natl Acad Sci USA 91:6083–6087
Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1 containing loci in the human lineage. Mol Biol Evol 18:926–935
Boumil RM, Lee JT (2000) Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet 10:2225–2232
Boyle L, Ballard SG, Ward DC (1990) Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci USA 87:7757–7761
Brockdorff N (1998) The role of Xist in X-inactivation. Curr Opin Genet Dev 8:328–333
Carrel L, Cottle AA, Goglin KC, Willard HF (1999) A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA 96:14440–14444
Claudio PP, Tonini T, Giordano A (2002) The retinoblastoma family: twins or distant cousins? Genome Biol 3:3012.1–3012.9
Cost GJ, Golding A, Schlissel MS, Boeke JD (2001) Target DNA chromatinization modulates nicking by L1 endonuclease. Nucl Acid Res 29:573–577
Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG (2003) Engineering evolution to study speciation in yeasts. Nature 422:68–72
Disteche CM (1999) Escapees on the X chromosome. Proc Natl Acad Sci USA 96:14180–14182
Dobigny G (2002) Spéciation chromosomique chez les espèces jumelles ouest-africaines du genre Taterillus (Rodentia, Gerbillinae): implications systématiques et biogéographiques, hypothèses génomiques. PhD thesis. Museum National d’Histoire Naturelle, Paris
Dobigny G, Aniskin V, Volobouev V (2002a) Explosive chromosomal evolution and speciation in the gerbil genus Taterillus (Rodentia, Gerbillinae): a case of two new cryptic species. Cytogenet Genome Res 96:117–124
Dobigny G, Baylac M, Denys C (2002b) Geometric morphometrics, neural networks and diagnosis of sibling Taterillus species (Rodentia, Gerbillinae). Biol J Linnean Soc 77:319–327
Dobigny G, Granjon L, Aniskin V, Bâ K, Volobouev V (2003a) A new sibling species of Taterillus (Rodentia, Gerbillinae) from West Africa. Mamm Biol 68:299–316
Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V (2003b) Evolution of rRNA genes clusters and telomeric evolution during explosive genome repatterning in Taterillus (Rodentia, Gerbillinae). Cytogenet Genome Res (in press)
Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N (1999) Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet 8:195–204
Eissenberg JC, Elgin SCR (2000) The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10:204–210
Fischer C, Ozouf-Costaz C, Roest Crollius H, Dasilva C, Jaillon O, Bouneau L, Bonillo C, Weissenbach J, Bernot A (2000) Karyotype and chromosomal location of characteristic tandem repeats in the pufferfish Tetraodon nigroviridis. Cytogenet Cell Genet 88:50–55
Gilbert DM (2002) Replication timing and transcriptional control: beyond cause and effects. Curr Opin Cell Biol 14:377–383
Goto T, Monk M (1998) Regulation of X-chromosome inactivation in development in mice and human. Microbiol Mol Biol Evol 62:362–378
Hansen RS (2003) X inactivation-specific methylation of LINE-1 elements by the DNMT3b: implications for the Lyon repeat hypothesis. Hum Mol Genet 12:2559–2567
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Kasahara S, Dutrillaux B (1983) Chromosome banding patterns of four species of bats, with special reference to a case of X-autosome translocation. Ann Genet 26:197–201
King M (1993) Species evolution: the role of chromosomal change. Cambridge University, Cambridge
Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120
Lifschytz E, Lindsley DL (1972) The role of X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci USA 69:182–189
Lin CC, Sasi R, Fan YS, Chen ZQ (1991) New evidence for tandem chromosome fusions in the karyotypic evolution of Asian muntjacs. Chromosoma 101:19–24
Lyon MF (1961) Gene action in the X chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Lyon MF (2000) LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci USA 97:6248–6249
Metcalfe CJ, Eldridge MCB, Toder R, Jonhston PG (1998) Mapping the distribution of the teloméric sequence (T2AG3) in the Macropodidae (Marsupilia) by fluorescence in situ hybridization. I. The swamp wallaby, Wallabia bicolor. Chromosome Res 6:603–610
Metzler-Guillemain C, Luciani J, Depetris D, Guichaoua MR, Mattei MG (2003) HP1b and HP1g, but not HP1a, decorate the entire XY body during human male meiosis. Chromosome Res 11:73–81
Mulligan G, Jacks T (1998) The retinoblastoma gene family: cousins with overlapping interests. Trends Genet 14:223–226
Neitzel H (1982) Karyotypenevolution und deren Bedeutung für den Speciationprozess der Cerviden (Cervidae, Artiodactyla, Mammalia). PhD thesis. University of Berlin
Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Caroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561–565
Parish DA, Vise P, Wichman HA, Bull JJ, Baker RJ (2002) Distribution of LINEs and other repetitive elements in the karyotype of the bat Carollia: implications for X-chromosome inactivation. Cytogenet Genome Res 96:191–197
Peters AHFM, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30:77–80
Quack B, Speed RM, Luciani JM, Noel B, Guichaoua M, Chandley AC (1988) Meiotic analysis of two human reciprocal X-autosome translocations. Cytogenet Cell Genet 48:43–47
Ratomponirina C, Viegas-Péquignot E, Dutrillaux B, Petter F, Rumpler Y (1986) Synaptonemal complexes in Gerbillidae: probable role of intercaled heterochromatin in gonosome–autosome translocations. Cytogenet Cell Genet 43:161–167
Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599
Ringrose L, Paro R (2001) Cycling silence. Nature 412:493–494
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972
Searle JB (1993) Chromosomal hybrid zones in Eutherian Mammals. In: Harrison H (ed) Hybrid zones and the evolutionary process. Oxford University, Oxford, pp 309–353
Sharp AJ, Spotswood HT, Robinson DO, Turner BM, Jacobs PA (2002) Molecular and cytogenetic analysis of the spreading of X inactivation in X:autosome translocations. Hum Mol Genet 11:3145–3146
Shi L, Yang F, Kumamoto A (1991) The chromosomes of tufted deer (Elaphodus cephalophus). Cytogenet Cell Genet 56:189–192
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Vassart T (1994) Evolution et diversité génétique chez les gazelles (Gazella): apports de l’électrophorèse des protéines, de la cytogénétique et des microsatellites. PhD thesis. Université de Paris XI, Orsay
Vaute O, Nicolas E, Vandel L, Trouche D (2002) Functional and physical interaction between the histone methyltransferase Suv39H1 and histone deacetylases. Nucl Acid Res 30:475–481
Vidal A, Koff A (2000) Cell-cycle inhibitors: three families united by a common cause. Gene 247:1–15
Viegas-Péquignot E, Dutrillaux B (1978) Une méthode simple pour obtenir des prophases et des prométaphases. Ann Genet 21:122–125
Viegas-Péquignot E, Benazzou T, Dutrillaux B, Petter F (1982) Complex evolution of sex chromosomes in Gerbillidae (Rodentia). Cytogenet Cell Genet 34:158–167
Volobouev V, Granjon L (1996) A finding of the XX/XY1Y2 sex-chromosome system in Taterillus arenarius (Gerbillinae, Rodentia) and its phylogenetic implications. Cytogenet Cell Genet 75:45–48
Watson JM, Spencer JA, Riggs AD, Graves JAM (1991) Sex chromosome evolution: Platypus gene mapping suggests that part of the human X chromosome was originally autosomal. Proc Natl Acad Sci USA 88:11256–11260
White MJD (1973) Animal cytology and evolution, 3rd edn. Cambridge University, Cambridge
White WM, Willard HF, Van Dyke DL, Wolffe DJ (1998) The spreading of X inactivation into autosomal material of an X-autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet 63:20–28
Yang F, O’Brien PCM, Wienberg J, Ferguson-Smith MA (1997) A reappraisal of the tandem fusion theory of karyotype evolution in the Indian muntjac using chromosome painting. Chromosome Res 5:109–117
Zhang P (1999) The cell cycle and development: redundant roles of cell cycle regulators. Curr Opin Cell Biol 11:655–662
Acknowledgements
We are grateful to J. Britton-Davidian, P.C.M. O’Brien, T.J. Robinson, K.A. Tolley and P. Waters for helpful comments on this manuscript, and to C. Canler, R. Cornette and J.P. Coutanceau for technical assistance. The suggestions made by four anonymous reviewers were greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Hassold
Rights and permissions
About this article
Cite this article
Dobigny, G., Ozouf-Costaz, C., Bonillo, C. et al. Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study. Chromosoma 113, 34–41 (2004). https://doi.org/10.1007/s00412-004-0292-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-004-0292-6