Abstract
Objective
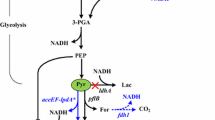
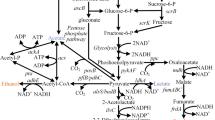
Metabolic engineering efforts are guided by identifying gene targets for overexpression and/or deletion. Isobutanol, a biofuel candidate, is biosynthesized using the valine biosynthesis pathway and enzymes of the Ehrlich pathway. Most reported studies for isobutanol production in Escherichia coli employ multicopy plasmids, an approach that suffers from disadvantages such as plasmid instability, increased metabolic burden, and use of antibiotics to maintain selection pressure. Cofactor imbalance is another issue that may limit production of isobutanol, as two enzymes of the pathway utilize NADPH as a cofactor.
Results
To address these issues, we constructed E. coli strains with chromosomally-integrated, codon-optimized isobutanol pathway genes (ilvGM, ilvC, kivd, adh) selected on the basis of their cofactor preferences. Genes involved in diverting pyruvate flux toward fermentation byproducts were deleted. Metabolite analyses of the constructed strains revealed extracellular accumulation of significant amounts of isobutyraldehyde, a pathway intermediate, and the overflow metabolites 2,3-butanediol and acetol.
Conclusions
These results demonstrate that the genetic modifications carried out led to activation of alternative pathways that diverted carbon flux toward formation of unwanted metabolites. The present study highlights how precursor metabolites can be metabolized through enzymatic routes that have not been considered important in previous studies due to the different strategies employed therein. The insights gained from the present study will allow rational genetic modification of host cells for production of metabolites of interest.





Similar content being viewed by others
References
Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74
Akita H, Nakashima N, Hoshino T (2015) Bacterial production of isobutanol without expensive reagents. Appl Microbiol Biotechnol 99:991–999
Albermann C, Trachtmann N, Sprenger GA (2010) A simple and reliable method to conduct and monitor expression cassette integration into the Escherichia coli chromosome. Biotechnol J 5:32–38
Altaras NE, Cameron DC (1999) Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl Environ Microbiol 65:1180–1185
Altaras NE, Cameron DC (2000) Enhanced production of (R)-1,2-propanediol by metabolically engineered Escherichia coli. Biotechnol Prog 16:940–946
Atsumi S, Liao JC (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19:414–419
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89
Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85:651–657
Baez A, Cho KM, Liao JC (2011) High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl Microbiol Biotechnol 90:1681–1690
Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MM, Arnold FH (2011) Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab Eng 13:345–352
Bennett GN, San KY (2001) Microbial formation, biotechnological production and applications of 1,2-propanediol. Appl Microbiol Biotechnol 55:1–9
Boronat A, Aguilar J (1979) Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol 140:320–326
Brautaset T, Lale R, Valla S (2009) Positively regulated bacterial expression systems. Microb Biotechnol 2:15–30
Brinkmann-Chen S, Cahn JK, Arnold FH (2014) Uncovering rare NADH-preferring ketol-acid reductoisomerases. Metab Eng 26:17–22
Brynildsen MP, Liao JC (2009) An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol 5:277
Cameron DC, Altaras NE, Hoffman ML, Shaw AJ (1998) Metabolic engineering of propanediol pathways. Biotechnol Prog 14:116–125
Chen YM, Lin EC (1984) Dual control of a common L-1,2-propanediol oxidoreductase by L-fucose and L-rhamnose in Escherichia coli. J Bacteriol 157:828–832
Cherepanov PP, Wackernagel W (1995) Gene disruption in Escherichia coli: tcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14
Chin JW, Khankal R, Monroe CA, Maranas CD, Cirino PC (2009) Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol Bioeng 102:209–220
Chunduru SK, Mrachko GT, Calvo KC (1989) Mechanism of ketol acid reductoisomerase–steady-state analysis and metal ion requirement. Biochemistry 28:486–493
Cooper RA (1984) Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol 38:49–68
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
de la Plaza M, Fernández de Palencia P, Peláez C, Requena T (2004) Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett 238:367–374
Deb SS, Reshamwala SMS, Lali AM (2016) A series of template plasmids for Escherichia coli genome engineering. J Microbiol Methods 125:49–57
Egan SM, Schleif RF (1993) A regulatory cascade in the induction of rhaBAD. J Mol Biol 234:87–98
Gedi V, Yoon MY (2012) Bacterial acetohydroxyacid synthase and its inhibitors–a summary of their structure, biological activity and current status. FEBS J 279:946–963
Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW (2006) Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 40:355–364
Habrych M, Rodriguez S, Stewart JD (2002) Purification and identification of an Escherichia coli beta-keto ester reductase as 2,5-diketo-d-gluconate reductase YqhE. Biotechnol Prog 18:257–261
Haldimann A, Daniels LL, Wanner BL (1998) Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol 180:1277–1286
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hill CM, Pang SS, Duggleby RG (1997) Purification of Escherichia coli acetohydroxyacid synthase isoenzyme II and reconstitution of active enzyme from its individual pure subunits. Biochem J 327:891–898
Huo YX, Cho KM, Rivera JG, Monte E, Shen CR, Yan Y, Liao JC (2011) Conversion of proteins into biofuels by engineering nitrogen flux. Nat Biotechnol 29:346–351
Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89:249–257
Kabus A, Georgi T, Wendisch VF, Bott M (2007) Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol 75:47–53
Ko J, Kim I, Yoo S, Min B, Kim K, Park C (2005) Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J Bacteriol 187:5782–5789
Kolodziej R, Scheib J (2012) Bio-isobutanol: the next-generation biofuel. hydrocarbon processing. http://www.hydrocarbonprocessing.com/magazine/2012/september-2012/special-report-refining-developments/bio-isobutanol-the-next-generation-biofuel. Accessed on 9 Dec 2018
Koo BM, Yoon MJ, Lee CR, Nam TW, Choe YJ, Jaffe H, Peterkofsky A, Seok YJ (2004) A novel fermentation/respiration switch protein regulated by enzyme IIAGlc in Escherichia coli. J Biol Chem 279:31613–31621
Lawther RP, Calhoun DH, Adams CW, Hauser CA, Gray J, Hatfield GW (1981) Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci USA 78:922–925
Lee C, Kim I, Lee J, Lee KL, Min B, Park C (2010) Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J Bacteriol 192:4205–4214
Lemuth K, Steuer K, Albermann C (2011) Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb Cell Fact 10:29
Liu Z, Liu P, Xiao D, Zhang X (2016) Improving isobutanol production in metabolically engineered Escherichia coli by co-producing ethanol and modulation of pentose phosphate pathway. J Ind Microbiol Biotechnol 43:851–860
Milne N, Wahl SA, van Maris AJA, Pronk JT, Daran JM (2016) Excessive by-product formation: a key contributor to low isobutanol yields of engineered Saccharomyces cerevisiae strains. Metab Eng Commun 3:39–51
Nielsen DR, Yoon SH, Yuan CJ, Prather KL (2010) Metabolic engineering of acetoin and meso-2, 3-butanediol biosynthesis in E. coli. Biotechnol J 5:274–284
Niu W, Guo J (2015) Stereospecific microbial conversion of lactic acid into 1,2-propanediol. ACS Synth Biol 4:378–382
Opel ML, Hatfield GW (2001) DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol Microbiol 39:191–198
Park JH, Lee KH, Kim TY, Lee SY (2007) Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA 104:7797–7802
Pick A, Rühmann B, Schmid J, Sieber V (2013) Novel CAD-like enzymes from Escherichia coli K-12 as additional tools in chemical production. Appl Microbiol Biotechnol 97:5815–5824
Rhee KY, Senear DF, Hatfield GW (1998) Activation of gene expression by a ligand-induced conformational change of a protein-DNA complex. J Biol Chem 273:11257–11266
Rhee KY, Opel M, Ito E, Hung SP, Arfin SM, Hatfield GW (1999) Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc Natl Acad Sci USA 96:14294–14299
Rodriguez GM, Atsumi S (2012) Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb Cell Fact 11:90
Rodriguez GM, Atsumi S (2014) Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237
Sabido A, Martínez LM, de Anda R, Martínez A, Bolívar F, Gosset G (2013) A novel plasmid vector designed for chromosomal gene integration and expression: use for developing a genetically stable Escherichia coli melanin production strain. Plasmid 69:16–23
Savrasova EA, Kivero AD, Shakulov RS, Stoynova NV (2011) Use of the valine biosynthetic pathway to convert glucose into isobutanol. J Ind Microbiol Biotechnol 38:1287–1294
Shafqat J, Höög JO, Hjelmqvist L, Oppermann UC, Ibáñez C, Jörnvall H (1999) An ethanol-inducible MDR ethanol dehydrogenase/acetaldehyde reductase in Escherichia coli: structural and enzymatic relationships to the eukaryotic protein forms. Eur J Biochem 263:305–311
Shi A, Zhu X, Lu J, Zhang X, Ma Y (2013) Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng 16:1–10
Silber P, Chung H, Gargiulo P, Schulz H (1974) Purification and properties of a diacetyl reductase from Escherichia coli. J Bacteriol 118:919–927
Tashiro Y, Rodriguez GM, Atsumi S (2015) 2-Keto acids based biosynthesis pathways for renewable fuels and chemicals. J Ind Microbiol Biotechnol 42:361–373
Tötemeyer S, Booth NA, Nichols WW, Dunbar B, Booth IR (1998) From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol 27:553–562
Trinh CT, Li J, Blanch HW, Clark DS (2011) Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl Environ Microbiol 77:4894–4904
Wek RC, Hatfield GW (1988) Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol 203:643–663
Westerfield WW (1945) A colorimetric determination of blood acetoin. J Biol Chem 161:495–502
Yan Y, Liao JC (2009) Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol 36:471–479
Yum DY, Lee BY, Pan JG (1999) Identification of the yqhE and yafB genes encoding two 2,5-diketo-D-gluconate reductases in Escherichia coli. Appl Environ Microbiol 65:3341–3346
Zhu Y, Lin EC (1989) L-1,2-propanediol exits more rapidly than L-lactaldehyde from Escherichia coli. J Bacteriol 171:862–867
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deb, S.S., Reshamwala, S.M.S. & Lali, A.M. Activation of alternative metabolic pathways diverts carbon flux away from isobutanol formation in an engineered Escherichia coli strain. Biotechnol Lett 41, 823–836 (2019). https://doi.org/10.1007/s10529-019-02683-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02683-5