Abstract
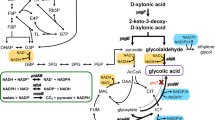
A critical factor in the biotechnological production of l-lysine with Corynebacterium glutamicum is the sufficient supply of NADPH. The membrane-integral nicotinamide nucleotide transhydrogenase PntAB of Escherichia coli can use the electrochemical proton gradient across the cytoplasmic membrane to drive the reduction of NADP+ via the oxidation of NADH. As C. glutamicum does not possess such an enzyme, we expressed the E. coli pntAB genes in the genetically defined C. glutamicum lysine-producing strain DM1730, resulting in membrane-associated transhydrogenase activity of 0.7 U/mg protein. When cultivated in minimal medium with 10% (w/v) carbon source, the presence of transhydrogenase slightly reduced glucose consumption, whereas the consumption of fructose, glucose plus fructose, and, in particular, sucrose was stimulated. Biomass was increased by pntAB expression between 10 and 30% on all carbon sources tested. Most importantly, the lysine concentration was increased in the presence of transhydrogenase by ∼10% on glucose, ∼70% on fructose, ∼50% on glucose plus fructose, and even by ∼300% on sucrose. Thus, the presence of a proton-coupled transhydrogenase was shown to be an efficient way to improve lysine production by C. glutamicum. In contrast, pntAB expression had a negative effect on growth and glutamate production of C. glutamicum wild type.

Similar content being viewed by others
References
Abe S, Takayama K, Kinoshita S (1967) Taxonomical studies on glutamic acid producing bacteria. J Gen Appl Microbiol 13:279–301
Anderlund M, Nissen TL, Nielsen J, Villadsen J, Rydstrom J, Hahn-Hagerdal B, Kielland-Brandt MC (1999) Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl Environ Microbiol 65:2333–2340
Ausubel F, Brent R, Kingston RE, Moore DD, Seidmann JG, Smith JA, Struhl K (1992) Preparation of genomic DNA. In: Ausubel F (ed) Short protocols in molecular biology. Wiley, New York, pp A1–A60
Bott M, Niebisch A (2003) The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104:129–153
Bott M, Niebisch A (2005) Respiratory energy metabolism. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC, Boca Raton, pp 307–334
Clarke DM, Bragg PD (1985) Cloning and expression of the transhydrogenase gene of Escherichia coli. J Bacteriol 162:367–373
Dominguez H, Lindley ND (1996) Complete sucrose metabolism requires fructose phosphotransferase activity in Corynebacterium glutamicum to ensure phosphorylation of liberated fructose. Appl Environ Microbiol 62:3878–3880
Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND (1998) Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem 254:96–102
Eggeling L (1994) Biology of l-lysine overproduction by Corynebacterium glutamicum. Amino Acids 6:261–272
Eggeling L, Sahm H (1999) l-glutamate and l-lysine: traditional products with impetuous developments. Appl Microbiol Biotechnol 52:146–153
Eikmanns BJ, Kleinertz E, Liebl W, Sahm H (1991) A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression and promoter probing. Gene 102:93–98
Eikmanns BJ, Rittmann D, Sahm H (1995) Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J Bacteriol 177:774–782
Georgi T, Rittmann D, Wendisch VF (2005) Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab Eng 7:291–301
Gourdon P, Baucher MF, Lindley ND, Guyonvarch A (2000) Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol 66:2981–2987
Hanahan D (1985) Techniques for transformation of E. coli. In: Glover DM (ed) DNA-cloning: a practical approach, IRL, Oxford, pp. 109–135
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Jackson JB (2003) Proton translocation by transhydrogenase. FEBS Lett 545: 18–24
Kelle R, Hermann T, Bathe B (2005) l-Lysine production. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC, Boca Raton, pp 467–490
Keilhauer C, Eggeling L, Sahm H (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 17:5595–5603
Kiefer P, Heinzle E, Zelder O, Wittmann C (2004) Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl Environ Microbiol 70:229–239
Kimura E (2005) l-Glutamate production. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC, Boca Raton, pp 441–465
Kotrba P, Inui M, Yukawa H (2001) The ptsI gene encoding enzyme I of the phosphotransferase system of Corynebacterium glutamicum. Biochem Biophys Res Commun 289:1307–1313
Liebl W (2005) Corynebacterium taxonomy. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC, Boca Raton, pp 9–34
Lindroth P, Mopper K (1979) High performance liquid chromatographic determination of subpicomole amounts of amino acids by pre-column fluorescence derivatization with o-phthaldialdehyde. Anal Chem 51:1667–1674
Marx A, de Graaf AA, Wiechert W, Eggeling L, Sahm H (1996) Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng 49:111–129
Marx A, Striegel K, de Graaf AA, Sahm H, Eggeling L (1997) Response of the central metabolism of Coyrnebacterium glutamicum to different flux burdens. Biotechnol Bioeng 56:168–180
Marx A, Eikmanns BJ, Sahm H, de Graaf AA, Eggeling L (1999) Response of the central metabolism in Corynebacterium glutamicum to the use of an NADH-dependent glutamate dehydrogenase. Metab Eng 1:35–48
Marx A, Hans S, Mockel B, Bathe B, de Graaf AA (2003) Metabolic phenotype of phosphoglucose isomerase mutants of Corynebacterium glutamicum. J Biotechnol 104:185–197
Moon MW, Kim HJ, Oh TK, Shin CS, Lee JS, Kim SJ, Lee JK (2005) Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett 244:259–266
Moritz B, Striegel K, de Graaf AA, Sahm H (2000) Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur J Biochem 267:3442–3452
Niebisch A, Bott M (2003) Purification of a cytochrome bc 1-aa 3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum. Identification of a fourth subunit of cytochrome aa 3 oxidase and mutational analysis of diheme cytochrome c 1. J Biol Chem 278:4339–4346
Ohnishi J, Mitsuhashi S, Hayashi M, Ando S, Yokoi H, Ochiai K, Ikeda M (2002) A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl Microbiol Biotechnol 58:217–223
Parche S, Burkovski A, Sprenger GA, Weil B, Kramer R, Titgemeyer F (2001) Corynebacterium glutamicum: a dissection of the PTS. J Mol Microbiol Biotechnol 3:423–428
Pfefferle W, Möckel B, Bathe B, Marx A (2003) Biotechnological manufacture of lysine. Adv Biochem Eng Biotechnol 79:59–112
Pons A, Dussap CG, Pequinot C, Gros JB (1996) Metabolic flux distribution in Corynebacterium melassecola ATCC 17965 for various carbon sources. Biotechnol Bioeng 51:77–189
Radmacher E, Stansen KC, Besra GS, Alderwick LJ, Maughan WN, Hollweg G, Sahm H, Wendisch VF, Eggeling L (2005) Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis, elicits l-glutamate efflux of Corynebacterium glutamicum. Microbiology 151:1359–1368
Rittmann D, Schaffer S, Wendisch VF, Sahm H (2003) Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch Microbiol 180:285–292
Sahm H, Eggeling L, de Graaf AA (2000) Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol Chem 381:899–910
Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
van der Rest ME, Lange C, Molenaar D (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogenic plasmid DNA. Appl Microbiol Biotechnol 52:541–545
Venning JD, Jackson JB (1999) A shift in the equilibrium constant at the catalytic site of proton-translocating transhydrogenase: significance for a ‘binding-change’ mechanism. Biochem J 341:329–337
Acknowledgement
Financial support by R&D Feed Additives of Degussa AG is gratefully acknowledged. C. glutamicum strain DM1730 was a kind gift of Dr. Brigitte Bathe, Degussa AG.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kabus, A., Georgi, T., Wendisch, V.F. et al. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol 75, 47–53 (2007). https://doi.org/10.1007/s00253-006-0804-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0804-9