Abstract
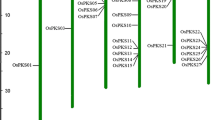
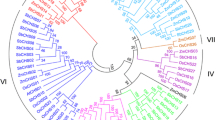
Systematic searches using the complete genome sequence of rice (Oryza sativa) identified OsSUS7, a new member of the rice sucrose synthase (OsSUS) gene family, which shows only nine single nucleotide substitutions in the OsSUS5 coding sequence. Comparative genomic analysis revealed that the synteny between OsSUS5 and OsSUS7 is conserved, and that significant numbers of transposable elements are scattered at both loci. In particular, a 17.6-kb genomic region containing transposable elements was identified in the 5′ upstream sequence of the OsSUS7 gene. GFP fusion experiments indicated that OsSUS5 and OsSUS7 are largely associated with the plasma membrane and partly with the cytosol in maize mesophyll protoplasts. RT-PCR analysis and transient expression assays revealed that OsSUS5 and OsSUS7 exhibit similar expression patterns in rice tissues, with the highest expression evident in roots. These results suggest that two redundant genes, OsSUS5 and OsSUS7, evolved via duplication of a chromosome region and through the transposition of transposable elements.
Similar content being viewed by others
References
Anguenot, R., Nguyen-Quoc, B., Yelle, S., and Michaud, D. (2006). Protein phosphorylation and membrane association of sucrose synthase in developing tomato fruit. Plant Physiol. Biochem. 44, 294–300.
Arai, M., Mori, H., and Imaseki, H. (1992). Expression of the gene for sucrose synthase during growth of mung bean seedlings. Plant Cell Physiol. 33, 503–506.
Balk, P.A., and de Boer, A.D. (1999). Rapid stalk elongation in tulip (Tulipa gesneriana L. cv. Apeldoorn) and the combined action of cold-induced invertase and the water-channel protein gammaTIP. Planta 209, 346–354.
Barratt, D.H., Barber, L., Kruger, N.J., Smith, A.M., Wang, T.L., and Martin, C. (2001). Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol. 127, 655–664.
Barratt, D.H., Derbyshire, P., Findlay, K., Pike, M., Wellner, N., Lunn, J., Feil, R., Simpson, C., Maule, A.J., and Smith, A.M. (2009). Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 106, 13124–13129.
Baud, S., Vaultier, M.N., and Rochat, C. (2004). Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 55, 397–409.
Bieniawska, Z., Paul Barratt, D.H., Garlick, A.P., Thole, V., Kruger, N.J., Martin, C., Zrenner, R., and Smith, A.M. (2007). Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 49, 810–828.
Carlson, S.J., Chourey, P.S., Helentjaris, T., and Datta, R. (2002). Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene. Plant Mol. Biol. 49, 15–29.
Chengappa, S., Guilleroux, M., Phillips, W., and Shields, R. (1999). Transgenic tomato plants with decreased sucrose synthase are unaltered in starch and sugar accumulation in the fruit. Plant Mol. Biol. 40, 213–221.
Cho, J.I., Lee, S.K., Ko, S., Kim, H.K., Jun, S.H., Lee, Y.H., Bhoo, S.H., Lee, K.W., An, G., Hahn, T.R., et al. (2005). Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep. 24, 225–236.
Cho, J.I., Ryoo, N., Ko, S., Lee, S.K., Lee, J., Jung, K.H., Lee, Y.H., Bhoo, S.H., Winderickx, J., An, G., et al. (2006). Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224, 598–611.
Cho, J.I., Ryoo, N., Eom, J.S., Lee, D.W., Kim, H.B., Jeong, S.W., Lee, Y.H., Kwon, Y.K., Cho, M.H., Bhoo, S.H., et al. (2009). Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759.
Cho, J.I., Burla, B., Lee, D.W., Ryoo, N., Hong, S.K., Kim, H.B., Eom, J.S., Choi, S.B., Cho, M.H., Bhoo, S.H., et al. (2010). Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol. 186, 657–668.
Chourey, P.S., Taliercio, E.W., Carlson, S.J., and Ruan, Y.L. (1998). Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 259, 88–96.
Craig, J., Barratt, P., Tatge, H., Dejardin, A., Handley, L., Gardner, C.D., Barber, L., Wang, T., Hedley, C., Martin, C., et al. (1999). Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J. 17, 353–362.
D’Aoust, M.A., Yelle, S., and Nguyen-Quoc, B. (1999). Antisense inhibition of tomato fruit sucrose synthase decreases fruit setting and the sucrose unloading capacity of young fruit. Plant Cell 11, 2407–2418.
Duncan, K.A., and Huber, S.C. (2007). Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Plant Cell Physiol. 48, 1612–1623.
Etxeberria, E., and Gonzalez, P. (2003). Evidence for a tonoplast-associated form of sucrose synthase and its potential involvement in sucrose mobilization from the vacuole. J. Exp. Bot. 54, 1407–1414.
Fallahi, H., Scofield, G.N., Badger, M.R., Chow, W.S., Furbank, R.T., and Ruan, Y.L. (2008). Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J. Exp. Bot. 59, 3283–3295.
Feng, Q., Zhang, Y., Hao, P., Wang, S., Fu, G., Huang, Y., Li, Y., Zhu, J., Liu, Y., Hu, X., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420, 316–320.
Fu, H., and Park, W.D. (1995). Sink- and vascular-associated sucrose synthase functions are encoded by different gene classes in potato. Plant Cell 7, 1369–1385.
Godt, D.E., Riegel, A., and Roitsch, T. (1995). Regulation of sucrose synthase expression in Chenopodium rubrum: characterization of sugar induced expression in photoautotrophic suspension cultures and sink tissue specific expression in plants. J. Plant Physiol. 146, 231–238.
Goff, S.A., Ricke, D., Lan, T.H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100.
Harada, T., Satoh, S., Yoshioka, T., and Ishizawa, K. (2005). Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Ann. Bot. (Lond) 96, 683–692.
Hesse, H., and Willmitzer, L. (1996). Expression analysis of a sucrose synthase gene from sugar beet (Beta vulgaris L.). Plant Mol. Biol. 30, 863–872.
Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788.
Hirose, T., Scofield, G.N., and Terao, T. (2008). An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174, 534–543.
Hohnjec, N., Becker, J.D., Puhler, A., Perlick, A.M., and Kuster, H. (1999). Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol. Gen. Genet. 261, 514–522.
Horst, I., Welham, T., Kelly, S., Kaneko, T., Sato, S., Tabata, S., Parniske, M., and Wang, T.L. (2007). TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 144, 806–820.
Huang, J.W., Chen, J.T., Yu, W.P., Shyur, L.F., Wang, A.Y., Sung, H.Y., Lee, P.D., and Su, J.C. (1996). Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci. Biotechnol. Biochem. 60, 233–239.
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389.
Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jiang, N., Feschotte, C., Zhang, X., and Wessler, S.R. (2004). Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr. Opin. Plant Biol. 7, 115–119.
Jun, S.H., Han, M.J., Lee, S., Seo, Y.S., Kim, W.T., and An, G. (2004). OsEIN2 is a positive component in ethylene signaling in rice. Plant Cell. Physiol. 45, 281–289.
Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195.
Karimi, M., Bleys, A., Vanderhaeghen, R., and Hilson, P. (2007). Building blocks for plant gene assembly. Plant Physiol. 145, 1183–1191.
Kim, J., Jun, S.H., Kang, H.G., Lee, J., and An, G. (2002). Molecular characterization of a GA-inducible gene, Cvsus1, in developing watermelon seeds. Mol. Cells 14, 255–260.
Kleines, M., Elster, R.C., Rodrigo, M.J., Blervacq, A.S., Salamini, F., and Bartels, D. (1999). Isolation and expression analysis of two stress-responsive sucrose-synthase genes from the resurrection plant Craterostigma plantagineum (Hochst.). Planta 209, 13–24.
Klotz, K.L., and Haagenson, D.M. (2008). Wounding, anoxia and cold induce sugarbeet sucrose synthase transcriptional changes that are unrelated to protein expression and activity. J. Plant Physiol. 165, 423–434.
Komatsu, A., Moriguchi, T., Koyama, K., Omura, M., and Akihama, T. (2002). Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J. Exp. Bot. 53, 61–71.
Komatsu, M., Shimamoto, K., and Kyozuka, J. (2003). Two-step regulation and continuous retrotransposition of the rice LINE-type retrotransposon Karma. Plant Cell 15, 1934–1944.
Kwon, S.J., Park, K.C., Son, J.H., Bureau, T., Park, C.H., and Kim, N.S. (2009). Sequence diversity of a domesticated transposase gene, MUG1, in Oryza species. Mol. Cells 30, 459–465.
Li, C.R., Zhang, X.B., Huang, C.H., and Hew, C.S. (2004). Cloning, characterization and tissue specific expression of a sucrose synthase gene from tropical epiphytic CAM orchid Mokara Yellow. J. Plant Physiol. 161, 87–94.
Ronald, P.C. (1998). Resistance gene evolution. Curr. Opin. Plant Biol. 1, 294–298.
Ruan, Y.L., Llewellyn, D.J., and Furbank, R.T. (2003). Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15, 952–964.
Salanoubat, M., and Belliard, G. (1987). Molecular cloning and sequencing of sucrose synthase cDNA from potato (Solanum tuberosum L.): preliminary characterization of sucrose synthase mRNA distribution. Gene 60, 47–56.
Sánchez de la Hoz, P., Vicente-Carbajosa, J., Mena, M., and Carbonero, P. (1992). Homologous sucrose synthase genes in barley (Hordeum vulgare) are located in chromosomes 7H (syn. 1) and 2H. Evidence for a gene translocation? FEBS Lett. 310, 46–50.
Sasaki, T., Matsumoto, T., Yamamoto, K., Sakata, K., Baba, T., Katayose, Y., Wu, J., Niimura, Y., Cheng, Z., Nagamura, Y., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420, 312–316.
Sebkova, V., Unger, C., Hardegger, M., and Sturm, A. (1995). Biochemical, physiological, and molecular characterization of sucrose synthase from Daucus carota. Plant Physiol. 108, 75–83.
Silvente, S., Camas, A., and Lara, M. (2003). Heterogeneity of sucrose synthase genes in bean (Phaseolus vulgaris L.): evidence for a nodule-enhanced sucrose synthase gene. J. Exp. Bot. 54, 749–755.
Sturm, A. (1999). Invertase. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 121, 1–7.
Sturm, A., and Tang, G.Q. (1999). The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 4, 401–407.
Sturm, A., Lienhard, S., Schatt, S., and Hardegger, M. (1999). Tissue-specific expression of two genes for sucrose synthase in carrot (Daucus carota L.). Plant Mol. Biol. 39, 349–360.
Subbaiah, C.C., Palaniappan, A., Duncan, K., Rhoads, D.M., Huber, S.C., and Sachs, M.M. (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J. Biol. Chem. 281, 15625–15635.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plantunique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741.
Uemura, T., Ueda, T., Ohniwa, R.L., Nakano, A., Takeyasu, K., and Sato, M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65.
van Ghelue, M., Ribeiro, A., Solheim, B., Akkermans, A.D., Bisseling, T., and Pawlowski, K. (1996). Sucrose synthase and enolase expression in actinorhizal nodules of Alnus glutinosa: comparison with legume nodules. Mol. Gen. Genet. 250, 437–446.
Wang, A.Y., Yu, W.P., Juang, R.H., Huang, J.W., Sung, H.Y., and Su, J.C. (1992). Presence of three rice sucrose synthase genes as revealed by cloning and sequencing of cDNA. Plant Mol. Biol. 18, 1191–1194.
Wang, F., Smith, A.G., and Brenner, M.L. (1993). Isolation and sequencing of tomato fruit sucrose synthase cDNA. Plant Physiol. 103, 1463–1464.
Werr, W., Frommer, W.B., Maas, C., and Starlinger, P. (1985). Structure of the sucrose synthase gene on chromosome 9 of Zea mays L. EMBO J. 4, 1373–1380.
Winter, H., Huber, J.L., and Huber, S.C. (1997). Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett. 420, 151–155.
Winter, H., Huber, J.L., and Huber, S.C. (1998). Identification of sucrose synthase as an actin-binding protein. FEBS Lett. 430, 205–208.
Yu, J., Hu, S., Wang, J., Wong, GK., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92.
Zrenner, R., Salanoubat, M., Willmitzer, L., and Sonnewald, U. (1995). Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 7, 97–107.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Cho, JI., Kim, HB., Kim, CY. et al. Identification and characterization of the duplicate rice sucrose synthase genes OsSUS5 and OsSUS7 which are associated with the plasma membrane. Mol Cells 31, 553–561 (2011). https://doi.org/10.1007/s10059-011-1038-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10059-011-1038-y