Abstract
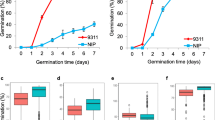
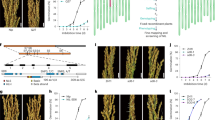
Seeds acquire primary dormancy during their development and the phytohormone abscisic acid (ABA) is known to play a role in inducing the dormancy. qSD12 is a major seed dormancy quantitative trait locus (QTL) identified from weedy rice. This research was conducted to identify qSD12 candidate genes, isolate the candidates from weedy rice, and determine the relation of the dormancy gene to ABA. A fine mapping experiment, followed by marker-assisted progeny testing for selected recombinants, narrowed down qSD12 to a genomic region of <75 kb, where there are nine predicted genes including a cluster of six transposon/retrotransposon protein genes and three putative (a PIL5, a hypothetic protein, and a bHLH transcription factor) genes based on the annotated Nipponbare genome sequence. The PIL5 and bHLH genes are more likely to be the QTL candidate genes. A bacterial artificial chromosome (BAC) library equivalent to 8–9 times of the haploid genome size was constructed for the weedy rice. One of the two BAC contigs developed from the library covers the PIL5 to bHLH interval. A pair of lines different only in the QTL-containing region of <200 kb was developed as isogenic lines for the qSD12 dormancy and non-dormancy alleles. The dormant line accumulated much higher ABA in 10-day developing seeds than the non-dormant line. In the QTL-containing region there is no predicted gene that has been assigned to ABA biosynthetic or metabolic pathways. Thus, it is concluded that the qSD12 underlying gene promotes ABA accumulation in early developing seeds to induce primary seed dormancy.


Similar content being viewed by others
References
Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103:17042–17047
Cai HW, Morishima H (2000) Genomic regions affecting seed shattering and seed dormancy in rice. Theor Appl Genet 100:840–846
Delouche JC, Burgos NR, Gealy DR, De San Martin GZ, Labrada R, Larinde M, Rosell C (2007) Weedy rices-origin, biology, ecology, and control. FAO Plant Production and Protection Paper 188, FAO, Rome
Destefano-Beltrán L, Knauber D, Huckle L, Suttle JC (2006) Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol Biol 61:687–697
Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, Zhang Y, Lu Q, Wang Y, Lu C, Han B, Chen F, Cheng Z, Chu C (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54:177–189
Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171:501–523
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415
Gao FY, Ren GJ, Lu XJ, Sun SX, Li HJ, Gao YM, Luo H, Yan WG, Zhang YZ (2008) QTL analysis for resistance to preharvest sprouting in rice (Oryza sativa). Plant Breed 127:268–273
Gao FY, Lu XJ, Wang WM, Sun SS, Li ZH, Li HJ, Ren GJ (2009) Trait-specific improvement of a cytoplasmic male-sterile line using molecular marker-assisted selection in rice. Crop Sci 49:99–106
Gianinetti A, Vernieri P (2007) On the role of abscisic acid in seed dormancy of red rice. J Exp Bot 58:3449–3462
Gu X-Y, Foley ME (2007) Epistatic interactions of three loci regulate flowering time under short and long daylengths in a backcross population of rice. Theor Appl Genet 114:745–754
Gu X-Y, Kianian SF, Foley ME (2004) Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166:1503–1516
Gu X-Y, Kianian SF, Foley ME (2005) Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice. Genetics 171:695–704
Gu X-Y, Kianian SF, Foley ME (2006) Dormancy genes from weedy rice respond divergently to seed development environments. Genetics 172:1199–1211
Gu X-Y, Turnipseed EB, Foley ME (2008) The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics 179:2263–2273
Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, afterripening, dormancy and germination. New Phytol 179:33–54
IRGSP (International Rice Genome Sequencing Project) (2005) The map-based sequence of the rice genome. Nature 436:793–800
Karssen CM, Brinkhorst-Van der Swan DLC, Breekland AE, Koornneef M (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157:158–165
Lee SJ, Oh CS, Suh JP, McCouch SR, Ahn SN (2005) Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed 124:209–219
Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45:309–319
Li C, Zhou A, Sang T (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170:185–193
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by phytochrome interacting factor 3-like5 during seed germination in Arabidopsis. Plant Cell 21:403–419
Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15:1998–2006
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
SAS Institute Inc. (1999) SAS/STAT user’s guide, version 8. Cary, NC
Song B-K, Nadarajah K, Romanov MN, Ratnam W (2005) Cross-species bacterial artificial chromosome (BAC) library screening via overgo-based hybridization and BAC-contig mapping of a yield enhancement quantitative trait locus (QTL) YLD1.1 in the Malaysian wild rice Oryza rofipogon. Cell Mol Biol Lett 10:425–437
Suh HS, Sato YI, Morishima H (1997) Genetic characterization of weedy rice (Oryza sativa L.) based on morpho-physiology, isozymes and RAPD markers. Theor Appl Genet 94:316–321
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. International Rice Research Institute, Manila
Acknowledgments
We thank B. Carsrud, Y. Wang and E. Castaneda for technical support and Dr. Carter for help with BAC library screening. Funding for this research was supported by grants from National Science Foundation (0641376) and in part from South Dakota Agriculture Extension Station (SD00H171-06IHG) and United States Department of Agriculture Cooperative State Research, Education, and Extension Service (SD00074-G).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gu, XY., Liu, T., Feng, J. et al. The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol 73, 97–104 (2010). https://doi.org/10.1007/s11103-009-9555-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9555-1