Abstract
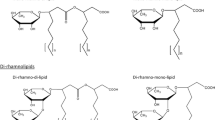
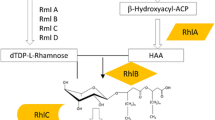
Rhamnolipids are biosurfactants consisting of rhamnose (Rha) molecules linked through a β-glycosidic bond to 3-hydroxyfatty acids with various chain lengths, and they have an enormous potential for various industrial applications. The best known native rhamnolipid producer is the human pathogen Pseudomonas aeruginosa, which produces short-chain rhamnolipids mainly consisting of a Rha-Rha-C10-C10 congener. Bacteria from the genus Burkholderia are also able to produce rhamnolipids, which are characterized by their long-chain 3-hydroxyfatty acids with a predominant Rha-Rha-C14-C14 congener. These long-chain rhamnolipids offer different physicochemical properties compared to their counterparts from P. aeruginosa making them very interesting to establish novel potential applications. However, widespread applications of rhamnolipids are still hampered by the pathogenicity of producer strains and—even more important—by the complexity of regulatory networks controlling rhamnolipid production, e.g., the so-called quorum sensing system. To overcome encountered challenges of the wild type, the responsible genes for rhamnolipid biosynthesis in Burkholderia glumae were heterologously expressed in the non-pathogenic Pseudomonas putida KT2440. Our results show that long-chain rhamnolipids from Burkholderia spec. can be produced in P. putida. Surprisingly, the heterologous expression of the genes rhlA and rhlB encoding an acyl- and a rhamnosyltransferase, respectively, resulted in the synthesis of two different mono-rhamnolipid species containing one or two 3-hydroxyfatty acid chains in equal amounts. Furthermore, mixed biosynthetic rhlAB operons with combined genes from different organisms were created to determine whether RhlA or RhlB is responsible to define the fatty acid chain lengths in rhamnolipids.



Similar content being viewed by others
References
Abdel-Mawgoud AM, Lépine F, Déziel E (2010) Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86(5):1323–1336. https://doi.org/10.1007/s00253-010-2498-2
Abdel-Mawgoud AM, Lépine F, Déziel E (2014) A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem Biol 21(1):156–164. https://doi.org/10.1016/j.chembiol.2013.11.010
Andrä J, Rademann J, Howe J, Koch MHJ, Heine H, Zähringer U, Brandenburg K (2006) Endotoxin-like properties of a rhamnolipid exotoxin from Burkholderia (Pseudomonas) plantarii: immune cell stimulation and biophysical characterization. Biol Chem 387(3):301–310. https://doi.org/10.1515/BC.2006.040
Banat I, Franzetti A, Gandolfi I, Bestetti G, Martinotti M, Fracchia L, Smyth T, Marchant R (2010) Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol 87(2):427–444. https://doi.org/10.1007/s00253-010-2589-0
Beuker J, Barth T, Steier A, Wittgens A, Rosenau F, Henkel M, Hausmann R (2016a) High titer heterologous rhamnolipid production. AMB Express 6(1):124. https://doi.org/10.1186/s13568-016-0298-5
Beuker J, Steier A, Wittgens A, Rosenau F, Henkel M, Hausmann R (2016b) Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor. AMB Express 6(1):11. https://doi.org/10.1186/s13568-016-0183-2
Chandrasekaran EV, Bemiller JN (1980) Constituent analysis of glycosaminoglycans. In: Methods in Carbohydrate chemistry, Vol. 3, Academic Press, New York, pp 89–96
Choi KH, Kumar A, Schweizer HP (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbial Methods 64(3):391–397. https://doi.org/10.1016/j.mimet.2005.06.001
Costa SGVAO, Déziel E, Lépine E (2011) Characterization of rhamnolipid production by Burkholderia glumae. Lett Appl Microbiol 53(6):620–627. https://doi.org/10.1111/j.1472-765X.2011.03154.x
de Lorenzo V, Eltis L, Kessler B, Timmis KN (1993) Analysis of Pseudomonas gene products using lacI q/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123(1):17–24. https://doi.org/10.1016/0378-1119(93)90533-9
Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R (1999) Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta 1440(2-3):244–252. https://doi.org/10.1016/S1388-1981(99)00129-8
Déziel E, Lépine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149(8):2005–2013. https://doi.org/10.1099/mic.0.26154-0
Dubeau D, Déziel E, Woods DE, Lépine F (2009) Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol 9(1):263–274. https://doi.org/10.1186/1471-2180-9-263
Elshikh M, Funston S, Ahmed S, Marchant R, Banat IB (2017) Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. New Biotechnol 36:26–36. https://doi.org/10.1016/j.nbt.2016.12.009
Funston SJ, Tsaousi K, Rudden M, Smyth TJ, Stevenson PS, Marchant R, Banat IM (2016) Characterising rhamnolipid production in Burkholderia thailandensis E264, a non-pathogenic producer. Appl Microbiol Biotechnol 100(18):7945–7956. https://doi.org/10.1007/s00253-016-7564-y
Giani C, Wullbrandt D, Rothert R, Meiwes J (1997) Pseudomonas aeruginosa and its use in a process for the biotechnological preparation of L-rhamnose. US005658793A. Hoechst AG, Frankfurt a. M
Grant SGN, Jessee J, Bloom FR, Hanahan D (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87(12):4645–4649. https://doi.org/10.1073/pnas.87.12.4645
Ham JH, Melanson RA, Rush MC (2011) Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12(4):329–339. https://doi.org/10.1111/j.1364-3703.2010.00676.x
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580. https://doi.org/10.1016/S0022-2836(83)80284-8
Hancock RE, Carey AM (1979) Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol 140(3):902–910
Häußler S, Nimtz M, Domke T, Wray V, Steinmetz I (1998) Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect Immun 66(4):1588–1593
Häußler S, Rohde M, von Neuhoff N, Nimtz M, Steinmetz I (2003) Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect Immun 71(5):2970–2975. https://doi.org/10.1128/IAI.71.5.2970-2975.2003
Hausmann S (2009) Einfluss des Lipase-spezifischen Chaperons LipH auf die Faltung und Sekretion der Lipasen LipA und LipC aus Pseudomonas aeruginosa. Dissertation, Heinrich-Heine-Universität Düsseldorf, Germany
Henkel M, Geissler M, Weggenmann F, Hausmann R (2017) Production of microbial biosurfactants: status quo of rhamnolipid and surfactin towards large-scale production. Biotechnol J 12(7). https://doi.org/10.1002/biot.201600561
Henkel M, Schmidberger A, Kühnert C, Beuker J, Bernard T, Schwartz T, Syldatk C, Hausmann R (2013) Kinetic modeling of the time course of N-butyryl-homoserine lactone concentration during batch cultivations of Pseudomonas aeruginosa PAO1. Appl Microbiol Biotechnol 97(17):7607–7616. https://doi.org/10.1007/s00253-013-5024-5
Hörmann B, Müller MM, Syldatk C, Hausmann R (2010) Rhamnolipid production by Burkholderia plantarii DSM 9509T. Eur J Lipid Sci Tech 112(6):674–680. https://doi.org/10.1002/ejlt.201000030
Jarvis FG, Johnson MJ (1949) A glycolipid produced by Pseudomonas aeruginosa. J Am Chem Soc 71(12):4124–4126. https://doi.org/10.1021/ja01180a073
Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, Hwang I (2003) Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis 87(8):890–895. https://doi.org/10.1094/PDIS.2003.87.8.890
Johann S, Seiler TB, Tiso T, Bluhm K, Blank LM, Hollert H (2016) Mechanism-specific and whole-organism ecotoxicity of mono-rhamnolipids. Sci Total Environ 548-549:155–163. https://doi.org/10.1016/j.scitotenv.2016.01.066
Lang S, Wullbrandt D (1999) Rhamnose lipids – biosynthesis, microbial production and application potential. Appl Microbiol Biotechnol 51(1):22–32. https://doi.org/10.1007/s002530051358
Lim J, Lee TH, Nahm BH, Choi YD, Kim M, Hwang I (2009) Complete genome sequence of Burkholderia glumae BGR1. J Bacteriol 191(11):3758–3759. https://doi.org/10.1128/JB.00349-09
Maier RM, Soberón-Chávez G (2000) Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol 54(5):625–633. https://doi.org/10.1007/s002530000443
Manso Pajarron A, de Koster CG, Heerma W, Schmidt M, Haverkamp J (1993) Structure identification of natural rhamnolipid mixtures by fast atom bombardment tandem mass spectrometry. Glycoconj J 10(3):219–226. https://doi.org/10.1007/BF00702203
Mata-Sandoval JC, Karns J, Torrents A (1999) HPLC method for characterization of rhamnolipids mixtures produced by Pseudomonas aeruginosa UG2 on corn oil. J Chromatogr 864(2):211–220. https://doi.org/10.1016/S0021-9673(99)00979-6
Müller MM, Hausmann R (2011) Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production. Appl Microbiol Biotechnol 91(2):251–264. https://doi.org/10.1007/s00253-011-3368-2
Müller MM, Hörmann B, Kugel M, Syldatk C, Hausmann R (2011) Evaluation of rhamnolipid production capacity of Pseudomonas aeruginosa PAO1 in comparison to the rhamnolipid over-producer strains DSM 7108 and DSM 2874. Appl Microbiol Biotechnol 89(3):585–592. https://doi.org/10.1007/s00253-010-2901-z
Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Chris Lee P, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen JA, Timmis KN, Düsterhöft A, Tümmler B, Fraser CM (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4(12):799–808. https://doi.org/10.1046/j.1462-2920.2002.00366.x
Nickzad A, Lépine F, Déziel E (2015) Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS One 10(6):e0128509. https://doi.org/10.1371/journal.pone.0128509
Nitschke M, Costa SGVAO, Contiero J (2005) Rhamnolipid surfactants: an update on the general aspects of these remarkable biomolecules. Biotechnol Prog 21(6):1593–1600. https://doi.org/10.1021/bp050239p
Ochsner UA (1993) Genetics and biochemistry of Pseudomonas aeruginosa rhamnolipid biosurfactant synthesis. Dissertation, ETH Zürich, Switzerland
Ochsner UA, Fiechter A, Reiser J (1994a) Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269(31):19787–19795
Ochsner UA, Koch AK, Fiechter A, Reiser J (1994b) Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol 176(7):2044–2054. https://doi.org/10.1128/jb.176.7.2044-2054.1994
Ochsner UA, Reiser J (1995) Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 92(14):6424–6428. https://doi.org/10.1073/pnas.92.14.6424
Ochsner UA, Reiser J, Fiechter A, Witholt B (1995) Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl Environ Microbiol 61(9):3503–3506
Olvera C, Goldberg JB, Sánchez R, Soberón-Chávez G (1999) The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett 179(1):85–90. https://doi.org/10.1111/j.1574-6968.1999.tb08712.x
Pearson JP, Pesci EC, Iglewski BH (1997) Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179(18):5756–5767. https://doi.org/10.1128/jb.179.18.5756-5767.1997
Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS (2000) Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146(11):2803–2814. https://doi.org/10.1099/00221287-146-11-2803
Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G (2001) Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol 40(3):708–718. https://doi.org/10.1046/j.1365-2958.2001.02420.x
Rehm BH, Mitsky TA, Steinbüchel A (2001) Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl Environ Microbiol 67(7):3102–3109. https://doi.org/10.1128/AEM.67.7.3102-3109.2001
Rosenau F, Isenhardt S, Gdynia A, Tielker D, Schmidt E, Tielen P, Schobert M, Jahn D, Wilhelm S, Jaeger KE (2010) Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa. FEMS Microbiol Lett 309:25–34. https://doi.org/10.1111/j.1574-6968.2010.02017.x
Rosenau F, Jaeger KE (2000) Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82(11):1023–1032. https://doi.org/10.1016/S0300-9084(00)01182-2
Sambrook J, Russell DW (2001) Molecular cloning, 3rd edn. Cold Spring Habor Laboratory Press, New York
Syldatk C, Lang S, Matulovic U, Wagner F (1985a) Production of four interfacial active rhamnolipids from n-alkanes or glycerol by resting cells of Pseudomonas species DSM 2874. Z Naturforsch C 40(1-2):61–67. https://doi.org/10.1515/znc-1985-1-213
Syldatk C, Lang S, Wagner F, Wray V, Witte L (1985b) Chemical and physical characterization of four interfacial-active rhamnolipids from Pseudomonas spec. DSM 2874 grown on n-alkanes. Z Naturforsch C 40(1-2):51–60. https://doi.org/10.1515/znc-1985-1-212
Tavares LFD, Silva PM, Junqueira M, Mariano DCO, Nogueira FCS, Domont GB, Freire DMG Neves BC (2012) Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl Microbiol Biotechnol 97(5):1909–1921. https://doi.org/10.1007/s00253-012-4454-9
Tiso T, Sabelhaus A, Behrens B, Wittgens A, Rosenau F, Hayen H, Blank LM (2016) Creating metabolic demand as an engineering strategy in Pseudomonas putida – rhamnolipid synthesis as an example. Metab Eng Commun 3:234–244. https://doi.org/10.1016/j.meteno.2016.08.002
Tiso T, Thies S, Müller M, Tsvetanova L, Carraresi L, Bröring S, Jaeger K-E, Blank LM (2017) Rhamnolipids—production, performance, and application, in consequences of microbial interactions with hydrocarbons, oils and lipids. In: Lee SY (ed) Production of fuels and chemicals. Springer-Verlag GmbH, Berlin
Toribio J, Escalante AE, Soberón-Chávez G (2010) Rhamnolipids: production in bacteria other than Pseudomonas aeruginosa. Eur J Lipid Sci Technol 112(10):1082–1087. https://doi.org/10.1002/ejlt.200900256
Urakami T, Ito-Yoshida C, Araki H, Kijima T, Suzuki KI, Komagata K (1994) Transfer of Pseudomonas plantarii and Pseudomonas glumae to Burkholderia as Burkholderia spp. and description of Burkholderia vandii sp. nov. Int J Syst Bacteriol 44(2):235–245. https://doi.org/10.1099/00207713-44-2-235
Verger R (1997) Interfacial activation of lipases: facts and artifacts. Trends Biotechnol 15(1):32–38. https://doi.org/10.1016/S0167-7799(96)10064-0
Vogel HJ, Bonner DM (1956) Acethylornithase of Escherichia coli: partial purification and some properties. J Biol Chem 218(1):97–106
Voget S, Knapp A, Poehlein A, Vollstedt C, Streit W, Daniel R, Jaeger KE (2015) Complete genome sequence of the lipase producing strain Burkholderia glumae PG1. J Biotechnol 204:3–4. https://doi.org/10.1016/j.jbiotec.2015.03.022
Wilhelm S, Gdynia A, Tielen P, Rosenau F, Jaeger KE (2007) The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol 189(18):6695–6703. https://doi.org/10.1128/JB.00023-07
Wittgens A, Kovacic F, Müller MM, Gerlitzki M, Santiago-Schübel B, Hofmann D, Tiso T, Blank LM, Henkel M, Hausmann R, Syldatk C, Wilhelm S, Rosenau F (2017) Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl Microbiol Biotechnol 101(7):2865–2878. https://doi.org/10.1007/s00253-016-8041-3
Wittgens A, Tiso T, Arndt TT, Wenk P, Hemmerich J, Müller C, Wichmann R, Küpper B, Zwick M, Wilhelm S, Hausmann R, Syldatk C, Rosenau F, Blank LM (2011) Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Factories 10(1):80. https://doi.org/10.1186/1475-2859-10-80
Zhang L, Veres-Schalnat TA, Somogyi A, Pemberton JE, Maier RM (2012) Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays. Appl Environ Microbiol 78(24):8611–8622. https://doi.org/10.1128/AEM.02111-12
Zhu K, Rock CO (2008) RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol 190(9):3147–3154. https://doi.org/10.1128/JB.00080-08
Acknowledgments
The authors are grateful to the Fachagentur Nachwachsende Rohstoffe e. V. (FNR) and the Deutsche Bundesstiftung Umwelt (DBU) for providing financial support.
Author information
Authors and Affiliations
Contributions
AW planned and executed the experiments, created figures, and drafted the manuscript; BSS and DH executed structural analysis of rhamnolipids and critically read the manuscript; MH and TT assisted the plasmid characterization and critically read the manuscript; LMB, RH, SW, and KEJ took part in initiating the research and critically read the manuscript; FR initiated the project, supervised the research, coordinated the study, and critically read the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest..
Electronic supplementary material
ESM 1
(PDF 537 kb)
Rights and permissions
About this article
Cite this article
Wittgens, A., Santiago-Schuebel, B., Henkel, M. et al. Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida—a step forward to tailor-made rhamnolipids. Appl Microbiol Biotechnol 102, 1229–1239 (2018). https://doi.org/10.1007/s00253-017-8702-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8702-x