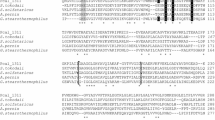
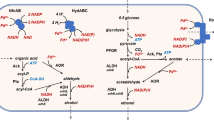
Abstract
Alcohol dehydrogenases are highly diverse enzymes catalysing the interconversion of alcohols and aldehydes or ketones. Due to their versatile specificities, these biocatalysts are of great interest for industrial applications. The adh3-gene encoding a group III alcohol dehydrogenase was isolated from the gram-positive bacterium Oenococcus oeni and was characterised after expression in the heterologous host Escherichia coli. Adh3 has been identified by genome BLASTP analyses using the amino acid sequence of 1,3-propanediol dehydrogenase DhaT from Klebsiella pneumoniae and group III alcohol dehydrogenases with known activity towards 1,3-propanediol as target sequences. The recombinant protein was purified in a two-step column chromatography approach. Crystal structure determination and biochemical characterisation confirmed that Adh3 forms a Ni2+-containing homodimer in its active form. Adh3 catalyses the interconversion of ethanol and its corresponding aldehyde acetaldyhyde and is also capable of using other alcoholic compounds as substrates, such as 1,3-propanediol, 1,2-propanediol and 1-propanol. In the presence of Ni2+, activity increases towards 1,3-propanediol and 1,2-propanediol. Adh3 is strictly dependent on NAD+/NADH, whereas no activity has been observed with NADP+/NADPH as co-factor. The enzyme exhibits a specific activity of 1.1 U/mg using EtOH as substrate with an optimal pH value of 9.0 for ethanol oxidation and 8.0 for aldehyde reduction. Moreover, Adh3 exhibits tolerance to several metal ions and organic solvents, but is completely inhibited in the presence of Zn2+. The present study demonstrates that O. oeni is a group III alcohol dehydrogenase with versatile substrate specificity, including Ni2+-dependent activity towards 1,3-propanediol.







Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Antoine E, Rolland JL, Raffin JP, Dietrich J (1999) Cloning and over-expression in Escherichia coli of the gene encoding NADPH group III alcohol dehydrogenase from Thermococcus hydrothermalis. Characterization and comparison of the native and the recombinant enzymes. Eur J Biochem 264:880–889
Ashok S, Raj SM, Rathnasingh C, Park S (2011) Development of recombinant Klebsiella pneumoniae dhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Appl Microbiol Biotechnol 90:1253–1265
Bennett GN, San KY (2001) Microbial formation, biotechnological production and applications of 1,2-propanediol. Appl Microbiol Biotechnol 55:1–9
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Britton HTK, Robinson RA (1931) CXCVIII.-Universal buffer solutions and the dissociation constant of veronal. J Chem Soc 458:1456–1462
Cameron DC, Altaras NE, Hoffman ML, Shaw AJ (1998) Metabolic engineering of propanediol pathways. Biotechnol Prog 14:116–125
Chatzifragkou A, Papanikolaou S, Dietz D, Doulgeraki AI, Nychas GJ, Zeng AP (2011) Production of 1,3-propanediol by Clostridium butyricum growing on biodiesel-derived crude glycerol through a non-sterilized fermentation process. Appl Microbiol Biotechnol 91:101–112
Chen R, Yin X, Yu C, Wang Q, Zhan X, Xie T (2011) Characterization of the recombinant group 3 alcohol dehydrogenase from Thermotoga lettingae TMO. Afr J of Microbiol Res 5:4004–4012
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D: Biol Crystallogr 66:12–21
Collaborative Computational Project 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D: Biol Crystallogr 50:760–763
Conway T, Sewell GW, Osman YA, Ingram LO (1987) Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol 169:2591–2597
Daniel R, Boenigk R, Gottschalk G (1995) Purification of 1,3-propanediol dehydrogenase from Citrobacter freundii and cloning, sequencing, and overexpression of the corresponding gene in Escherichia coli. J Bacteriol 177:2151–2156
Evans PR (1997) SCALA. Joint CCP4 and ESF-EAMBC. Newslon Protein Crystallogr 33:22–24
Fenghuan W, Huijin Q, He H, Tan T (2005) High-level expression of the 1,3-propanediol oxidoreductase from Klebsiella pneumoniae in Escherichia coli. Mol Biotechnol 31:211–219
Hensgens CMH, Jansen M, Nienhuis-Kuiper ME, Boekema EJ, Van Breemen JFL, Hansen TA (1995) Purification and characterization of an alcohol dehydrogenase from 1,2-propanediol-grown Desulfovibrio strain HDv. Arch Microbiol 164:265–270
Hernandez-Tobias A, Julian-Sanchez A, Pina E, Riveros-Rosas H (2011) Natural alcohol exposure: is ethanol the main substrate for alcohol dehydrogenases in animals? Chem Biol Interact 191:14–25
Horn U, Strittmatter W, Krebber A, Knupfer U, Kujau M, Wenderoth R, Muller K, Matzku S, Pluckthun A, Riesenberg D (1996) High volumetric yields of functional dimeric miniantibodies in Escherichia coli, using an optimized expression vector and high-cell-density fermentation under non-limited growth conditions. Appl Microbiol Biotechnol 46:524–532
Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89:249–257
Johnson EA, Lin EC (1987) Klebsiella pneumoniae 1,3-propanediol:NAD+ oxidoreductase. J Bacteriol 169:2050–2054
Kabsch W (2010) Xds. Acta Crystallogr D: Biol Crystallogr 66:125–132
Kosjek B, Stampfer W, Pogorevc M, Goessler W, Faber K, Kroutil W (2004) Purification and characterization of a chemotolerant alcohol dehydrogenase applicable to coupled redox reactions. Biotechnol Bioeng 86:55–62
Luers F, Seyfried M, Daniel R, Gottschalk G (1997) Glycerol conversion to 1,3-propanediol by Clostridium pasteurianum: cloning and expression of the gene encoding 1,3-propanediol dehydrogenase. FEMS Microbiol Lett 154:337–345
Ma C, Zhang L, Dai J, Xiu Z (2010a) Relaxing the coenzyme specificity of 1,3-propanediol oxidoreductase from Klebsiella pneumoniae by rational design. J Biotechnol 146:173–178
Ma Z, Rao Z, Zhuge B, Fang H, Liao X, Zhuge J (2010b) Construction of a novel expression system in Klebsiella pneumoniae and its application for 1,3-propanediol production. Appl Biochem Biotechnol 162:399–407
Malaoui H, Marczak R (2000) Purification and characterization of the 1,3-propanediol dehydrogenase of Clostridium butyricum E5. Enzyme Microb Technol 27:399–405
Marcal D, Rego AT, Carrondo MA, Enguita FJ (2009) 1,3-propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity. J Bacteriol 191:1143–1151
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674
Meng F, Xu Y (2010) Purification and characterization of an anti-Prelog alcohol dehydrogenase from Oenococcus oeni that reduces 2-octanone to (R)-2-octanol. Biotechnol Lett 32:533–537
Mills DA, Rawsthorne H, Parker C, Tamir D, Makarova K (2005) Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol Rev 29:465–475
Montella C, Bellsolell L, Perez-Luque R, Badia J, Baldoma L, Coll M, Aguilar J (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J Bacteriol 187:4957–4966
Moon JH, Lee HJ, Park SY, Song JM, Park MY, Park HM, Sun J, Park JH, Kim BY, Kim JS (2011) Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD + cofactor. J Mol Biol 407:413–424
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Qi X, Guo Q, Wei Y, Xu H, Huang R (2012) Enhancement of pH stability and activity of glycerol dehydratase from Klebsiella pneumoniae by rational design. Biotechnol Lett 34:339–346
Rujanonon R, Praertsan P, Phongdara A, Panrat T, Sun J, Rappert S, Zeng AP (2011) Construction of recombinant E. coli expressing fusion protein to produce 1,3-propanediol. Int J of Biol and Med Sci 1:26–32
Sambrook J, Fritsch E, Maniatis T (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Skraly FA, Lytle BL, Cameron DC (1998) Construction and characterization of a 1,3-propanediol operon. Appl Environ Microbiol 64:98–105
Stevens MJ, Vollenweider S, Meile L, Lacroix C (2011) 1,3-propanediol dehydrogenases in Lactobacillus reuteri: impact on central metabolism and 3-hydroxypropionaldehyde production. Microb Cell Fact 10:61
Sulzenbacher G, Alvarez K, Van Den Heuvel RH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342:489–502
Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ (1990) Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD oxidoreductase. Appl Environ Microbiol 56:943–948
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Timpson LM, Liliensiek AK, Alsafadi D, Cassidy J, Sharkey MA, Liddell S, Allers T, Paradisi F (2013) A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl Microbiol Biotechnol 97:195–203
Veiga-da-Cunha M, Foster MA (1992) 1,3-propanediol:NAD+ oxidoreductases of Lactobacillus brevis and Lactobacillus buchneri. Appl Environ Microbiol 58:2005–2010
Walter KA, Bennett GN, Papoutsakis ET (1992) Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J Bacteriol 174:7149–7158
Wang F, Qu H, Zhang D, Tian P, Tan T (2007) Production of 1,3-propanediol from glycerol by recombinant E. coli using incompatible plasmids system. Mol Biotechnol 37:112–119
Xiu ZL, Zeng AP (2008) Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl Microbiol Biotechnol 78:917–926
Ying X, Grunden AM, Nie L, Adams MW, Ma K (2009) Molecular characterization of the recombinant iron-containing alcohol dehydrogenase from the hyperthermophilic Archaeon, Thermococcus strain ES1. Extremophiles 13:299–311
Acknowledgments
The authors thank Henning Piascheck for his help with the fermentation experiments. X-ray data collection was performed on the ID23-2 beamline at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. We are grateful to Cristoph Müller-Dickmann at ESRF for providing assistance in using beamline ID23-2. This work was funded by the Excellence Cluster in the Excellence Initative by the State of Hamburg “Fundamentals of Synthetic Biological Systems (SynBio)”. The work was also supported by the Hungarian National Scientific Research Foundation (OTKA) grant PD104365.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 404 kb)
Rights and permissions
About this article
Cite this article
Elleuche, S., Fodor, K., Klippel, B. et al. Structural and biochemical characterisation of a NAD+-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications. Appl Microbiol Biotechnol 97, 8963–8975 (2013). https://doi.org/10.1007/s00253-013-4725-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4725-0