Abstract
The major histocompatibility complex (MHC) plays a key role in immune defense, and the Mhc genes of cynomolgus macaque display a high degree of polymorphism. Based on their geographic distribution, different populations of cynomolgus macaques are recognized. Here we present the characterization of the Mhc class I and II repertoire of a large pedigreed group of cynomolgus macaques originating from the mainland north of the isthmus of Kra (N = 42). Segregation analyses resulted in the definition of 81 unreported Mafa-A/B/DRB/DQ/DP haplotypes, which include 32 previously unknown DRB regions. In addition, we report 13 newly defined Mafa-A/B/DRB/DQ/DP haplotypes in a group of cynomolgus macaques originating from the mainland south of the isthmus of Kra/Maritime Southeast Asia (N = 16). A relatively high level of sharing of Mafa-A (51%) and Mafa-B (40%) lineage groups is observed between the populations native to the north and the south of isthmus of Kra. At the allelic level, however, the Mafa-A/B haplotypes seem to be characteristic of a population. An overall comparison of all currently known data revealed that each geographic population has its own specific combinations of Mhc class I and II haplotypes. This illustrates the dynamic evolution of the cynomolgus macaque Mhc region, which was most likely generated by recombination and maintained by selection due to the differential pathogenic pressures encountered in different geographic areas.
Similar content being viewed by others
Introduction
Cynomolgus macaques (Macaca fascicularis, Mafa) belong to the group of Old World monkeys, and their natural habitat ranges from the mainland to the islands of Southeast Asia. In addition, a population of isolated cynomolgus macaques lives on the island of Mauritius near Eastern Africa and was introduced there approximately 400 years ago by Dutch/Portuguese sailors (Sussman and Tattersall 1986). The founder animals most likely originate from Java or Sumatra (Kawamoto et al. 2008; Tosi and Coke 2007). Cynomolgus macaques, also referred to as crab-eating or long-tailed macaques, share with humans a common ancestor that lived approximately 25–33 million years ago (Glazko and Nei 2003; Perelman et al. 2011). Both species share a highly similar immune system, and cynomolgus macaques are therefore often applied as animal model in biomedical research for various human diseases, such as AIDS, tuberculosis, dengue, SARS-CoV-2, zika, Parkinson’s and Huntington’s disease, as well as in studies for transplantation research (Almond et al. 2019; Boszormenyi et al. 2021; Breitbach et al. 2019; Dijkman et al. 2019; Emborg 2017; Kwon et al. 2019).
The major histocompatibility complex (MHC) is a large genomic region, mostly occupied by genes that encode molecules that play a central role in generating innate and adaptive immune responses. Some of them are cell surface structures. In humans, the region is located on chromosome 6 and is referred to as the human leucocyte antigen (HLA) system. One distinguishes class I and II molecules, in which the classical HLA class I molecules are designated as HLA-A, HLA-B, and HLA-C, whereas the class II molecules are referred to as HLA-DP, HLA-DQ, and HLA-DR. Orthologs of these molecules can be found in cynomolgus macaques and are named Mafa-A, Mafa-B, Mafa-DP, Mafa-DQ, and Mafa-DR. The ortholog of HLA-C is absent in macaques (Boyson et al. 1996). The Mhc region in humans is by far the most intensively studied. The use of cynomolgus macaques as models in biomedical research, however, has intensified the research into their Mhc cluster over the past 10–15 years, resulting in substantial information on its organization and its repertoire (Budde et al. 2010; Campbell et al. 2009; Creager et al. 2011; Doxiadis et al. 2006, 2010, 2012; Karl et al. 2017; Krebs et al. 2005; O’Connor et al. 2007; Otting et al. 2012, 2017; Pendley et al. 2008; Saito et al. 2012; Sano et al. 2006; Shiina et al. 2015; Shortreed et al. 2020; van der Wiel et al. 2015; Westbrook et al. 2015).
A hallmark of the Mhc is its extensive allelic polymorphism, observed for most of the class I and II genes. Moreover, some of the sections in the region are subject to expansion and contraction, which may result in copy number variation. Both of these characteristics give rise to a very dynamic region, reflected by variation between different species as well as by species-specific diversification. In humans, an Mhc class I haplotype (defined as a combination of genes inherited together on a chromosome) comprises a single copy of the classical HLA-A, HLA-B, and HLA-C genes. In contrast, in cynomolgus macaques, several Mafa-A and Mafa-B genes can be present on a haplotype, and the number of genes may vary substantially between individuals. The different genes might display differential levels of transcription, and those with high transcription levels are referred to as majors, whereas the lowly transcribed genes are referred to as minors (Otting 2005). For the Mafa-A region, each haplotype seems to contain at least one highly transcribed Mafa-A1 gene (major) (de Groot et al. 2020; Otting et al. 2005; Shortreed et al. 2020). Occasionally, the presence of a second Mafa-A1 gene on a haplotype, encoding a different lineage, may occur. In addition, a haplotype can encode several A genes that are transcribed at a lower level (minors), and these have been designated as Mafa-A2 to Mafa-A6 and Mafa-A8. The Mafa-B region, which is even more complex, can encode several B genes with a high transcription profile (majors), and haplotypes with up to seven major B genes have been described. However, the presence of one to three major B genes per haplotype is predominant (Budde et al. 2010; Saito et al. 2012; Shiina et al. 2015; Shortreed et al. 2020). On top of that, the region may comprise several B genes showing a low transcription profile (minors). The cynomolgus macaque Mhc genes show extensive polymorphism, and at present, 592 Mafa-A (of which 358 are Mafa-A1) and 1000 Mafa-B alleles have been reported (release version 3.7.0.0 IPD-MHC NHP Database).
Mhc class II gene products comprise an alpha and a beta polypeptide chain and are encoded by different genes: namely, DPA, DPB, DQA, DQB, DRA, and DRB. In humans and cynomolgus macaques, one DPA1/DPB1 and one DQA1/DQB1 gene tandem are located on a haplotype, as is a single DRA gene. Equivalents of the HLA-DQA2 and HLA-DQB2 genes are absent in macaques, and the DPA2 and DPB2 genes are considered pseudogenes in humans and macaques (Bontrop et al. 1999). The number of DRB genes can vary, and one to four genes/pseudogenes (excluding the DRB9 segment) per haplotype are documented in humans. In cynomolgus macaques, more extensive DRB copy number variation is reported, resulting in the presence of a high number of region configurations, which may contain two to six genes/pseudogenes (excluding the DRB9 segment) (Doxiadis et al. 2010, 2012). In humans, substantial levels of polymorphism are encountered for the DPB1, DQA1, DQB1, and DRB genes, whereas the DPA1 and DRA genes are oligomorphic (Robinson et al. 2020). For cynomolgus macaques, polymorphism is reported for all abovementioned Mhc class II genes, and this is mostly located in the exons encoding the peptide-binding cleft (Blancher et al. 2014; Creager et al. 2011; Doxiadis et al. 2006, 2012; Ling et al. 2011; Otting et al. 2012, 2017; Sano et al. 2006; van der Wiel et al. 2015). The Mafa-DRA gene forms an exception. For this gene, most nucleotide polymorphisms are synonymous, and those mutations that are non-synonymous are mainly located in the leader sequence.
The Biomedical Primate Research Centre (BPRC) houses a large pedigreed breeding colony of cynomolgus macaques that initially started with animals originating from the Indonesian islands, from the mainland south of the isthmus of Kra (continental Malaysia), and from Mauritius (Doxiadis et al. 2010). For this original cohort of animals, the Mhc class I and II repertoires have been studied in detail (Otting et al. 2012). Over the past decade, 84 cynomolgus macaques, originating from other breeding facilities, have been introduced into the original colony. Mitochondrial 12S rRNA gene segment analyses showed that some of these introduced animals are natively originating from the mainland south of the isthmus of Kra or Maritime Southeast Asia; however, most are characterized as originating from the mainland north of the isthmus of Kra. For this communication, we have thoroughly characterized the Mhc class I (Mafa-A and Mafa-B) and II (Mafa-DPA1/DPB1, Mafa-DQA1/DQB1, Mafa-DRB) repertoires of 58 of the recently introduced animals and their offspring, using a Pacific Biosciences (PacBio) sequencing protocol. In addition to the description of unreported Mhc class I and II alleles, this cohort study revealed unreported Mafa-A, Mafa-B, and Mafa-DRB haplotypes and Mafa-DQA1/DQB1 and Mafa-DPA1/DPB1 combinations, which resulted in the identification of 94 novel Mafa-A/B/DRB/DQ/DP haplotypes. Although sharing on a lineage and an allelic level is observed to some extent between cynomolgus macaques of different geographic areas, this large cohort study together with previously published data show that the different populations apparently have their own characteristic Mafa-A/B/DRB/DQ/DP configurations.
Materials and methods
Samples and geographic origin
Between 2011 and 2016, a large number of cynomolgus macaques (N = 84), originating from Chinese and other European breeding facilities, were introduced into the existing breeding colony housed at the BPRC, the Netherlands (also referred to as BPRC’s original breeding cohort). The original geographic origin of these animals was defined by a phylogenetic comparison of the mitochondrial 12S rRNA gene segments (Doxiadis et al. 2010). From this cohort, the Mhc class I and II repertoire was analyzed in depth. Offspring of these animals were included to deduce the Mhc haplotypes. Twenty-two of the 84 animals, however, did not produce offspring, which prevented us from deducing their Mhc haplotypes. In addition, for 4 of the 84 animals, both Mhc haplotypes were found to be identical to those present in the pre-existing cohort (Otting et al. 2012), and this data is not included. For the remainder 58 animals, the Mhc class I and II haplotypes are provided. For deducing these haplotypes, in total, 309 animals were analyzed (Table S1).
Mitochondrial 12S rRNA gene segment and phylogenetic analysis
Amplification of part of the mitochondrial 12S rRNA gene was performed essentially as described previously (Kocher et al. 1989), using a specific primer set (Table S2). The PCR for each sample was performed in a 50-μl reaction mixture containing 200–500 ng of mitochondrial DNA isolated from fresh EDTA blood or serum, 1 × PCR buffer, 2.5 mM MgCl2, 0.25 mM dNTP, and 5 U Taq polymerase (Thermo Fisher Scientific). The cycling reaction started with an initial denaturation step of 2 min at 95 °C, followed by 35 cycles each consisting of 20 s at 95 °C, 20 s at 55 °C, 40 s at 72 °C, and a final extension of 5 min at 72 °C. The PCR products were purified using the GeneJet Gel extraction kit (Thermo Fisher Scientific) and sequenced either on a ABI 3100, 3130, or 3500 genetic analyzer (Applied Biosystems, Foster City, USA) in accordance with the manufacturer’s guidelines. The data were analyzed using the Sequence Navigator program (Applied Biosystems), MacVector, or Geneious Prime 2021.1.1 software (Kearse et al. 2012). The eight unreported sequences (Table S3) were confirmed by identification in different animals and/or by doing two independent PCR reactions and submitted to the European Nucleotide Archive (accession numbers: OV260054, OV260064, OV260068, OV260184, OV260833, OV260835, OV260836, OV260461). Phylogenetic analysis was conducted with MEGA version 7.0.18 using the maximum likelihood method based on the Jukes-Cantor model (Tamura et al. 2004). The bootstrap values were inferred from 1000 replicates.
Genomic DNA isolation and microsatellite typing of STR-MHC-A (D6S2854 and D6S2859) and STR-MHC-DRB (D6S2878)
Genomic DNA (gDNA) was extracted from EDTA whole blood samples using a standard salting out procedure, or from ± 15 × 106 PBMCs with an AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Amplification of the relevant DNA segments in cynomolgus macaques was performed as described for rhesus macaques using the same primer sets (Doxiadis et al. 2007, 2011). For each sample, the PCR fragments of the STR-MHC-A microsatellite markers D6S2854 and D6S2859 were multiplexed in a 25-μl mixture. The multiplexed STR-MHC-A fragments and the STR-MHC-DRB (D6S2878) fragments were each mixed with size standards and separated on an automated capillary electrophoresis system (ABI 3500, Applies Biosystems). Fragment-length analyses were performed with Genemapper software (Applied Biosystems).
RNA isolation and MHC class I and II amplification
Total RNA was extracted directly from EDTA whole blood samples or from ± 15 × 106 PBMCs with the AllPrep DNA/RNA Mini Kit. First-strand complementary DNA (cDNA) was synthesized with the RevertAid First Strand cDNA Synthesis Kit (Invitrogen) in accordance with the manufacturer’s instructions.
Full-length Mhc class I (Mafa-A and Mafa-B) transcripts were obtained by amplification of total cDNA with a mixture of two forward and three reverse primers (Table S2), in accordance with a previously reported protocol (van der Wiel et al. 2018). For the amplification of full-length DRB (cDRB) (de Groot 2004), DPA1, DQA1, and DQB1 transcripts and partial DRB (~ 800 bp containing exon 2 and intron 2) at the genomic level (gDRB), a set of one forward and one reverse primer was used for each different application (Table S2). The partial DRB (gDRB) amplicon contains exon 2 and the STR-MHC-DRB, which allows us to characterize the STR-length coupled to each different DRB allele. Full-length DPB1 transcripts were amplified using a set of one forward and two reverse primers (Table S2). Each primer was tagged at the 5′ end with a unique 16-bp barcode (www.pacb.com) to allow identification of pooled samples after PacBio sequencing. The PCRs for the cDRB, DPA1, DPB1, DQA1, and DQB1 samples were performed in a 50-μl reaction mixture containing 5 μl of cDNA, 1 × Phusion HF buffer, 0.2 mM dNTPs, 0.4 μM of the differentially barcoded forward and reverse primer(s), 3% DMSO, and 0.02 U/μl Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific). The amplification started with an initial denaturation of 3 min at 98 °C, followed by 25 cycles, each consisting of 5 s at 98 °C, 10 s at 60 °C (for cDRB) or 10 s at 56 °C (for DPA1, DPB1, DQA1, and DQB1), and 20 s at 72 °C and a final extension of 5 min at 72 °C. For the amplification of gDRB, the same protocol was used as described for cDRB, except that 10 μl of gDNA (50 ng/μl) was added to the PCR mixture, and the amplification was performed with 30 cycles.
PCR products were size-selected (Table S2) by gel-electrophoresis and purified using the GeneJet Gel extraction kit. The DNA concentrations of the single samples were measured using the Qubit dsDNA HS assay kit and Qubit 2.0 Fluorometer (Thermo Fisher Scientific). The Mhc class I and II amplicons were pooled proportionately to their concentrations and number of expected alleles. For Mafa-A/B, Mafa-DRB, and Mafa-DQ/DP, respectively, 50, 40, and 15 ng per sample were used. The pooled samples were purified twice using AMPure XP beads (Beckman Coulter) at a 1:1 bead to DNA volume ratio, and the concentration was measured again and had to be more than 1 μg of total DNA. PacBio SMRTbell libraries were generated according to Pacific Biosciences “Procedure & Checklist — Amplicon Template Preparation and Sequencing,” and sequencing was performed by the Leiden Genome Technology Center using a Pacific Biosciences RSII (P4-C2 sequencing chemistry), Sequel I (P6-C4 sequencing chemistry), or Sequel II system (sequencing kit versions 2.0 and 2.1). Sequence data collection was performed with a 4, 10, or 20–24 h movie time to obtain sufficient yields of high-quality circular consensus reads, respectively.
PacBio data analysis
Circular consensus sequences (CCS) were selected for high read quality (value of 0.99 or higher) and demultiplexed based on unique barcoding. Geneious Prime 2021.1.1 software (Kearse et al. 2012) was used to map the CCS reads to a reference database, consisting of reported cynomolgus macaque Mhc class I or II sequences, to identify 100% matching reads (100% overlap, 0% mismatch, maximum ambiguity = 1). The unused reads were grouped and were de novo assembled. The consensus of each de novo contig was trimmed for the primer sequence and phylogenetically aligned with the cynomolgus macaque Mhc class I or II reference database. The novel sequences were confirmed by identification in two independent Pacbio runs and showing to segregate into families. Eight novel sequences (Mafa-A1*003:10, Mafa-A1*063:03:03, Mafa-B*077:04, Mafa-DRB*W026:02:02, Mafa-DQA1*24:09, Mafa-DQB1*28:02, Mafa-DPA1*02:55, and Mafa-DPB1*17:03:02) were detected in a single animal and confirmed by identification in two independent Pacbio runs taking a cutoff read number > 50 (100% identical). The novel sequences are submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) and IPD-MHC NHP Database (https://www.ebi.ac.uk/ipd/mhc/group/NHP/) (Maccari et al. 2017). For the definition of a Mhc haplotype, the focus was on the highly transcribed alleles (corresponding to a relatively high number of reads). The allele with the highest number of reads was defined as the “major 1” (Table S1).
Nomenclature
In the past, the characterization of the Mhc class II genes was mainly based on sequencing only exon 2, which is by far the most polymorphic exon for these types of genes. Recently, however, a large number of full-length macaque Mhc class II transcript sequences were archived in the IPD-MHC NHP Database. Phylogenetic analysis of all currently known macaque DQB1 sequences showed that several of the previously described alleles of the DQB1*17 and *18 lineages cluster into separate groups, and renaming has been proposed (Otting et al. 2017). The IPD-MHC NHP nomenclature committee introduced the renaming at the beginning of 2020, and three new lineages for the macaque DQB1 gene were introduced: namely, DQB1*26, DQB1*27, and DQB1*28 (https://www.ebi.ac.uk/ipd/mhc/group/NHP/). Consequently, particular alleles of the DQB1*17 and *18 lineages were renamed, and an overview of the renamed alleles relevant to the present study is provided (Table 1).
Results
Determining the geographic origin of the cynomolgus macaques using mitochondrial DNA analysis
The mitochondrial 12S rRNA gene segment, shown to be highly informative to elucidate the geographic origin of different macaques (de Groot et al. 2008; Doxiadis et al. 2010; Tosi et al. 2003), was sequenced for the 84 cynomolgus macaques introduced into the existing breeding colony of the BPRC. A phylogenetic comparison with reference data revealed that most of the animals, 61 in total, have a segment that clusters with reference sequences of cynomolgus macaques with an origin from the mainland north of the isthmus of Kra, 2 with reference sequences indicative of animals living on the mainland south of the isthmus of Kra, and 21 with reference sequences that match Indonesian and Malaysian island populations (Fig. 1; Table S3). In contrast, most of the animals of BPRC’s original breeding colony originate from the mainland south of the isthmus of Kra and the Indonesian and Malaysian islands and a few from the island of Mauritius (Doxiadis et al. 2010). In the remaining sections, the animals originating from the mainland north of the isthmus of Kra will be referred to as mainland N.i.K., whereas the animals originating from the mainland south of the isthmus of Kra and Indonesian and Malaysian islands are seen as one cohort and will be referred to as mainland S.i.K./Maritime Southeast Asia.
Phylogenetic analysis of the 12S rRNA segment of representative cynomolgus macaques from the 84 animals introduced into the BPRC breeding colony in comparison to reference sequences of cynomolgus macaques of known geographic origin (Table S3; Doxiadis et al. 2010). The 12S rRNA segments of rhesus macaques (Mamu) from Burmese, Chinese, and Indian origin are taken as an outgroup. Bootstrap values < 50 have been omitted. The brackets indicate the geographic clusters. The eight different 12S rRNA segments that were previously unreported are incorporated in phylogenetic analysis and recognizable by Ji-numbers. The map of the Asian continent depicted on the right side of the figure illustrates the natural habitat of cynomolgus macaques, with a division into the mainland area north of the isthmus of Kra and the mainland south of the isthmus of Kra and Indonesian and Malaysian islands. With regard to these two areas, the number of animals introduced into the original BPRC breeding colony is indicated. The island of Mauritius is located over 4000 miles off the southeast coast of Africa and is not illustrated
Microsatellite typing for STR-MHC-A and STR-MHC-DRB revealed new length patterns in the cynomolgus macaques
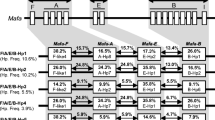
The microsatellite markers D6S2854 and D6S2859 for Mhc-A and D6S2878 for Mhc-DRB are shown to be highly polymorphic. Genotyping with these markers in rhesus and cynomolgus macaque has revealed different patterns that may consist of several peaks of variable length (de Groot et al. 2008; Doxiadis et al. 2011; Otting et al. 2012). The length patterns of these three microsatellite markers were previously shown to segregate with Mafa-A and Mafa-DRB haplotypes in the population of cynomolgus macaques housed at the BPRC, respectively (Otting et al. 2012). We determined the length patterns of D6S2854/D6S2859 and D6S2878 in the cohort of 84 cynomolgus macaques that had been introduced into the BPRC breeding colony. Here we report the STR typing of 58 introduced animals, for which one or more offspring were also analyzed to establish segregation of the STR markers (Table S1). In this panel, we describe 53 different STR patterns for the microsatellite markers D6S2854/D6S2859 and 63 distinct patterns for D6S2878 (Table S1). Three of the patterns were identical to ones observed in the cynomolgus macaques from BPRC’s original breeding cohort (Otting et al. 2012), and these were all Mhc-A STR patterns (Table S1, indicated by a gray background). Four of the identified Mhc-A STR patterns are shared between the cohort of animals originating from mainland N.i.K. and mainland S.iK/Maritime Southeast Asia (Table S1, D6S2854/D6S2859 patterns presented in boldface). None of the reported Mhc-DRB STR patterns was shared between the animals from these two well-defined populations. These results show that between the cynomolgus macaque populations originating from mainland N.i.K. and from mainland S.i.K/Maritime Southeast Asia/Mauritius a small percentage (5%) of Mhc-A STR patterns is shared, whereas these populations share no currently known Mhc-DRB STR patterns (Fig. 2) (this study) (Otting et al. 2012).
Venn diagrams illustrating the degree of sharing of Mhc-A (D6S2854 and D6S2859) and Mhc-DRB (D6S2878) STR patterns between cynomolgus macaques originating from mainland N.i.K. (black circle), mainland S.i.K./Maritime Southeast Asia (dark gray circle), and BPRC’s original cohort of cynomolgus macaques (light gray circle)
Mafa-A and Mafa-B polymorphism in cynomolgus macaques recently introduced in BPRC breeding cohort
Microsatellite typing resulted in the characterization of several distinct, previously unknown, Mhc-A STR patterns, for which the Mafa-A haplotypes that segregate with the patterns have been determined. In addition, the Mafa-B haplotypes linked to the different Mafa-A haplotypes have been characterized. Therefore, genetic material of 58 introduced animals and/or their offspring was sequenced using a PacBio sequencing platform. This resulted in the identification of 56 and 103 different major Mafa-A1 and Mafa-B alleles, respectively, and three alleles of the Mafa-A2*01 and Mafa-A2*24 lineages, which also show a high transcription profile and are considered to be majors (Table S1). In addition, alleles of the lower transcribed Mafa-A2*05, Mafa-A3*13, Mafa-A4*01/*14, Mafa-A5*30, and Mafa-A6*01 lineages as well as several lower transcribed Mafa-B alleles per animal were also identified. Segregation analyses showed that duplications are encountered on some haplotypes, and next to the major Mafa-A1 lineage member, a second copy (belonging to another lineage) was observed, which can be highly or lowly transcribed. For example, on the haplotype that contains Mafa-A1*018:01:01, the minor Mafa-A1*086:01:02 is encountered (Table S1, haplotype nos. 28–31), and Mafa-A1*056:03:01 is found together with Mafa-A1*090:03, which is considered a major as well (Table S1, haplotype no. 62). Most Mafa-A allele combinations are linked to a specific D6S2854/D6S2859 pattern, but some exceptions have been found. For instance, allele combination Mafa-A1*001:02:01/A2*05:57:01 is linked to two different STR patterns in animals originating from mainland N.i.K. (Table S1, haplotype nos. 2–4).
Of the majors, only two Mafa-A1 alleles and 11 Mafa-B alleles were also detected in animals from the original breeding colony (Table S1, alleles indicated with a salmon background) (Otting et al. 2012). Therefore, the introduced cynomolgus macaques extend the overall Mhc-A and Mhc-B repertoire of the BPRC breeding colony (Fig. 3, cohorts represented by black and dark gray circles), thereby expanding its outbred status. Most of the Mafa-A and Mafa-B alleles, encountered in the present study, represent known entities that are archived in the IPD-MHC NHP Database (Maccari et al. 2017). Five Mafa-A1, one Mafa-A2*05, and nine Mafa-B alleles were not previously reported (Table 2; Table S1). Between the two cohorts analyzed in this communication (mainland N.i.K. and mainland S.i.K./Maritime Southeast Asia), one major A and six B alleles are shared (Fig. 3; Table S1 alleles indicated with a $ sign). In addition, these cohorts share minors as well, such as Mafa-A4*01:02, Mafa-B*060:01, and Mafa-B*060:04 (Table S1, alleles indicated with an “&” sign). In animals originating from mainland N.i.K., the majors Mafa-A1*007:01 and Mafa-B*056:01:01 were most frequently observed (Table S1).
Venn diagrams illustrating the degree of sharing of the Mhc class I (major Mafa-A and Mafa-B) and II (Mafa-DRB, Mafa-DQA, Mafa-DQB, Mafa-DPA, and Mafa-DPB) alleles detected in the studied cohorts of cynomolgus macaques originating from mainland N.i.K. (black circle), mainland S.i.K/Maritime Southeast Asia (dark gray circle), and BPRC’s original cohort of cynomolgus macaques (light gray circle)
Mafa-DRB polymorphism linked to STR-MHC-DRB
To characterize the Mafa-DRB allelic repertoire that is linked to the 63 identified STR-MHC-DRB patterns, a stretch of 710–928 nucleotides, including exon 2 of Mhc-DRB and the adjacent microsatellite (D6S2878) located in intron 2, was sequenced at the gDNA level (gDRB). Most of the STR-MHC-DRB patterns are linked to a specific combination of Mafa-DRB alleles. As reported previously, we observed that one Mafa-DRB allele can be linked to different STR lengths. This seems to depend on the DRB haplotype it segregates on and is in agreement with the fact that STRs evolve faster than their adjacent coding sequence (de Groot et al. 2008; Doxiadis et al. 2007). For example, Mafa-DRB1*03:07 is found in conjunction with STR lengths 209, 220, and 224, segregating on haplotype nos. 56, 67, and 77 in animals originating from mainland N.i.K. and haplotype nos. 82 and 83 in animals originating from mainland S.i.K./Maritime Southeast Asia (Table S1). For haplotype nos. 67 and 77, the combination of DRB alleles is identical, but the different STR length patterns obviously refer to the distinct families they segregate in. For haplotype nos. 56 and 82–83, Mafa-DRB1*03:07 is found in combination with alleles of different lineages resulting in distinct region configuration.
In addition, we sequenced the Mafa-DRB alleles at the cDNA level, and in the panel of 58 animals, we characterized 76 full-length transcripts (Table S1, alleles indicated in bold). Three DRB1, two DRB*W, and the DBR6 (pseudogene) lineage alleles did not amplify at cDNA level, and characterization of these alleles was only at the gDNA level (comprising exon 2 and adjacent DRB-STR). Twenty-four alleles are shared with the DRB repertoire present in animals from the original BPRC cohort (Fig. 3; Table S1) (Otting et al. 2012). All DRB alleles detected were previously archived in the IPD-MHC NHP Database. The sequence length of two DRB alleles was extended (Table 2; Table S1). Five DRB alleles are shared between the two recently studied cohorts (Fig. 3; Table S1). The haplotypes Mafa-DRB1*03:03/-DRB*W001:01/-DRB*W003:02/-DRB6*01:28 (11 times) and Mafa-DRB1*04:13:01/-DRB*W037:02/-DRB6*01:13:01 (12 times) were most frequently observed in animals originating from mainland N.i.K (Table S1).
Characterization of the DQ and DP repertoire
In the present panel of 58 animals, 34 DQA1, 32 DQB1, 32 DPA1, and 30 DPB1 alleles were recovered. Seven alleles (one DQA1, four DPA1, and two DPB1) were newly identified, which were all detected in animals originating from mainland N.i.K., and for five alleles, the sequences were extended (Table 2). The majority of the DQ and DP alleles detected represent polymorphisms that are known and archived in the IPD-MHC NHP Database. Seven DQA1, seven DQB1, two DPA1, and eight DPB1 alleles are shared with alleles present in animals from BPRC’s original colony (Fig. 3; Table S1) (Otting et al. 2012). Between the two cohorts studied for this communication, six alleles (one DQA1, four DQB1, and one DPB1) are shared (Fig. 3; Table S1), whereas no sharing of DPA1 alleles was observed. As compared to Mafa-A, Mafa-B, and Mafa-DRB, characterization of the Mafa-DQ and Mafa-DP repertoire by deep sequencing revealed several alternatively spliced transcripts for the different genes (data not shown). An inventory showed that, in particular, Mafa-DQB is subject to alternative splicing, including for instance the skipping of exon 4, which codes for the transmembrane region. The generation of such an isoform is also predicted on the basis of certain HLA-DQB transcripts (Briata et al. 1989). The conserved character of this specific splice event between humans and cynomolgus macaques suggests biological relevance. Nevertheless, additional research is required to establish the exact impact of alternative splicing in Mhc class II genes in primates.
Strong linkage is documented for the DQA1/DQB1 and DPA1/DPB1 gene tandems, and a comprehensive overview of the recorded combinations has been provided, comparing two populations of Indonesian cynomolgus macaques with a population of cynomolgus macaques of Cambodian/Vietnamese origin (Otting et al. 2017). This latter population has a more extensive number of DQ and DP haplotypes in comparison to the two Indonesian cynomolgus macaque populations, and only some of the haplotypes are shared between the populations of different origin. In the animals analyzed for this study, most of the found DQ and DP combinations substantiate previously published data. Nine DQ and twelve DP combinations are new, with the majority found in animals originating from mainland N.i.K. (Fig. 4). The newly described combination Mafa-DQA1*01:14:04/-DQB*06:23 was observed in both cohorts (Fig. 4). This DQ pair was found in linkage with the DP pairs Mafa-DPA1*11:01/-DPB1*16:01 and Mafa-DPA1*09:02/-DPB1*17:01:02 (Table S1). These two DP pairs are also both documented in literature in animals originating from mainland N.i.K. and S.i.K./Maritime Southeast Asia (Otting et al. 2017). Based on the latest nomenclature update (see “materials and methods section” section), an overview is provided of the DQA1/DQB1 and DPA1/DPB1 lineage combinations that can be encountered in cynomolgus macaques (Fig. 5) (this study) (Otting et al. 2017; Shortreed et al. 2020).
Mafa-DQ and Mafa-DP pairs characterized in the two cohorts studied. Newly described Mafa-DQ and Mafa-DP pairs are indicated by a gray background. The number (number sign) of Mhc haplotypes on which a particular DQ or DP combination is found is provided (see also Table S1). In the last two columns, an “x” marks whether a DQ or DP combination was previously published in cynomolgus macaques originating either from Cambodian/Vietnam (Ca/Vi) and/or from the mainland south of the isthmus of Kra/Indonesian and Malaysian islands (Indo) (Otting et al. 2017; Shortreed et al. 2020). For three haplotypes, the DQA1 or DPA1 allele could not be determined (Table S1). The newly described combination Mafa-DQA1*01:14:04/-DQB*06:23 observed in both cohorts is indicated in boldface
Combinations of cynomolgus macaque DQA1/DQB1 and DPA1/DPB1 lineages observed in the present and previously characterized cohorts (indicated by black boxes). The shaded (Otting et al. 2017) and gray (Shortreed et al. 2020) boxes indicate combinations that were additionally described in previously characterized panels. For DQB1 lineages, the previously used designations are given in brackets (see “materials and methods section”) (Otting et al. 2017)
Cynomolgus macaque Mhc haplotypes
The segregation analysis of the Mafa class I and II repertoire in the cohort of animals characterized for this communication resulted in the identification of 94 different Mafa-A/B/DRB/DQ/DP haplotypes distinct from the ones identified in BPRC’s original breeding cohort (Otting et al. 2012). The analysis includes 81 haplotypes in animals originating from mainland N.i.K. and 13 in animals from mainland S.i.K./Maritime Southeast Asia (Table S1). Please note that the exact order of some of these genes is unknown at present.
In the panel of animals originating from mainland N.i.K. (N = 42), we identified 49 Mafa-A, 48 Mafa-B, and 35 Mafa-DRB haplotypes (Table S1), of which 23 Mafa-A and 27 Mafa-B haplotypes are newly identified (Fig. 6). The remaining Mafa-A and Mafa-B haplotypes were found in a panel of cynomolgus macaques that were of Vietnamese, Cambodian, and Cambodian/Indonesian mixed origin (Karl et al. 2017). A few of the newly characterized haplotypes show variation only at the allelic level when compared to haplotypes found in the panel of cynomolgus macaques of Vietnamese, Cambodian, and Cambodian/Indonesian mixed origin (Karl et al. 2017). For instance, Mafa-A1*003:07/-A4*14:03:01 differs for the minor A4 allele, Mafa-B*001:01:01/*044:14/*085:01:01/*030:03:01 for the Mafa-B*044 allele, and for haplotypes that contain a Mafa-B*028-lineage member; allelic variation is observed for either the Mafa-B*021, Mafa-B*030, Mafa-B*068, or Mafa-B*124-lineage allele. Furthermore, two Mafa-B haplotypes (Mafa-B*001:01:01/B*007:01:01 and Mafa-B*039:01:01/B*007:08:01) are presented as new in this study because they differ from the ones characterized in the panel of Karl and colleagues (2017) by lacking a B*030-lineage member.
Mafa-A/-B and Mafa-A/-B/-DRB haplotypes newly identified in cynomolgus macaques native to the mainland north of the isthmus of Kra and native to the mainland south of the isthmus of Kra/Maritime Southeast Asia, respectively. The major Mafa-A and Mafa-B alleles as well as the Mafa-DRB transcripts are printed in boldface with a dark gray background. Newly identified alleles (new) together with alleles for which the sequence information is extended (ext) are indicated. Allele resolution was sometimes not reached, in which case only the lineage designation is indicated. The haplotype numbers (Hapl.no.) on which the indicated Mafa-A, Mafa-B, and Mafa-DRB haplotypes segregate are shown and correspond to those indicated in Table S1. In the allele name, the suffix N indicates that the allele is characterized by a premature stop codon. N.d. stands for not determined
To the best of our knowledge, no data is currently available in the literature on DRB haplotypes in cynomolgus macaques native to mainland N.i.K. Therefore, the majority of the DRB haplotypes (32 of the 35) that we report are novel (Fig. 7, upper panel). The three DRB haplotypes in the lower panel of Fig. 7 had been reported earlier, but a precise indication of the geographic origin of the animals these haplotypes were detected in was lacking (Doxiadis et al. 2010). Furthermore, three of the haplotypes are also reported in cynomolgus macaques native to mainland S.i.K./Maritime Southeast Asia (Fig. 7, haplotypes within the black bordered boxes) (Otting et al. 2012; Shortreed et al. 2020).
Mafa-DRB haplotypes characterized in animals native to the mainland north of the isthmus of Kra. The transcribed DRB alleles are printed in boldface with a dark gray background. The allele for which the sequence information is extended (ext) is indicated. Allele resolution was sometimes not reached, in which case only the lineage designation is indicated. The haplotype numbers (Hapl.no.) on which the indicated DRB haplotypes segregate are shown, and they correspond to those indicated in Table S1. The black-bordered boxes mark the haplotypes that are also found in cynomolgus macaques from mainland S.i.K./Maritime Southeast Asia. ^The DRB1 allele for this haplotype could not be amplified at the cDNA level, and at the gDNA level, only exon 2 was sequenced, which does not distinguish between Mafa-DRB1*03:12:01 and Mafa-DRB1*03:36
The haplotypes that are most frequently observed in the cohort native to mainland N.i.K. are Mafa-A1*007:01/A2*05:22/A4*14:03:01 (6%), Mafa-B*039:01:01/B*007:01:01 (6%), Mafa-B*145:01:01/B*068:06:01/B*144:01:01 (7%), Mafa-DRB1*03:03/DRB*W001:01/DRB*W003:02 (13%), and Mafa-DRB1*04:13:01/W037:02 (14%) (Table S1). Most of the 42 analyzed animals of this cohort appear to differ for their Mafa-A/B/DRB/DQ/DP haplotypes. The sharing of a Mafa-A/B/DRB/DQ/DP haplotype was found in the case of five couples of cynomolgus macaques (Table S1, haplotype nos. 13, 29, 34, 48, 77), and mitochondrial 12S rRNA analysis suggests that some of these animals might be related. Two animals share both Mhc haplotypes (haplotype nos. 19 and 50) as well as their mitochondrial 12S rRNA gene segment, suggesting that they descended from the same parents.
In the group of animals native to mainland S.i.K./Maritime Southeast Asia, 11 of the 16 animals have either one Mafa-A/B/DRB/DQ/DP haplotype that is shared with a haplotype of the original breeding colony or have one haplotype that could not be confirmed in the contemporary offspring population. The 13 newly described Mafa-A/B/DRB/DQ/DP haplotypes consist of nine Mafa-A, seven Mafa-B, and seven Mafa-DRB haplotypes (Table S1), of which four, six, and three represent novel entities, respectively (Fig. 6) (Otting et al. 2012; Shortreed et al. 2020). The majority of these novel haplotypes show variation only at the allelic level. Haplotypes Mafa-A1*022:02/A4*01:02, Mafa-A1*032:01/A1*047:02/A4*01:09, Mafa-B*095:01:01/B*033:01:01, and Mafa-DRB1*03:07/DRB*W001:07/DRB*W026:02:01 seem to represent newly reported combinations. One of the haplotypes identified in the mainland S.i.K./Maritime Southeast Asia cohort is identical to the M2 haplotype defined in Mauritius cynomolgus macaques (Table S1, haplotype no. 91) (Wiseman et al. 2013).
Evidence for cross-over events
Within the cohort of animals studied for this communication, four different cross-over events did turn up (Fig. 8; Table S4). All of them were observed in offspring of the introduced animals native to mainland N.i.K. One of the recombination events occurred between the Mafa-A and Mafa-B regions and three between the Mafa-B and Mafa-DRB regions. In total, 283 individuals (representing introduced animals native to mainland N.i.K and their offspring) were analyzed, which yielded a frequency of recombination of 1.0% for the cross-over events observed between the Mafa-B and Mafa-DRB regions and 0.3% for the other event. These numbers are in line with earlier published data (Aarnink et al. 2014; Blancher et al. 2012a; Shortreed et al. 2020), and further support that in cynomolgus macaques, there may exist one or more recombination hot spots in the area located between the Mafa class I and II regions (de Groot et al. 2014).
Schematic presentation of the four cross-over events observed in the Mhc region in the offspring of four introduced animals originating from the mainland north of the isthmus of Kra. The Mafa-A, Mafa-B, and Mafa-DRB haplotypes are shown with their short names (Table S4). The putative cross-over position is indicated with a large x. The haplotype resulting from the cross-over event is illustrated below the arrow
Dynamics in cynomolgus macaque Mhc class I and II haplotypes
Cynomolgus macaques of Vietnamese/Cambodian, Filipino, Indonesian, and Mauritian origin have been intensively studied for their Mhc repertoire (Blancher et al. 2012b, 2014; Budde et al. 2010; Campbell et al. 2009; Creager et al. 2011; Doxiadis et al. 2012; Huang et al. 2019; Karl et al. 2017; Li et al. 2012; Ling et al. 2011, 2012; Otting et al. 2012, 2017; Sano et al. 2006; Shiina et al. 2015; Shortreed et al. 2020; van der Wiel et al. 2015; Zhang et al. 2012; Wang et al. 2011), and for some of the studied populations, Mafa-A/B or Mafa-A/B/DRB/DQ/DP haplotypes have been published (Budde et al. 2010; Karl et al. 2017; Otting et al. 2012; Saito et al. 2012; Shortreed et al. 2020). Here, we distinguish the different Mafa-A and Mafa-B haplotypes by means of the most abundant transcribed mRNA allele encoded by a particular haplotype (referred to as “major 1” in Table S1, and possibly referred to by others as diagnostic Mafa-A and Mafa-B) (Karl et al. 2017; Shortreed et al. 2020). The sequences of the “major 1” alleles cluster into lineages of which the majority are old entities and predate the speciation of macaques. A comparison analysis of all currently characterized Mafa-A/-B haplotypes indicates that particular “major 1” Mafa-A and Mafa-B lineage groups might only be present in cynomolgus macaques from a specific geographic region (Table S5). Of the 72 major Mafa-A and 62 major Mafa-B lineage groups identified, 37 (51%) and 25 (40%), respectively, are shared between populations native to mainland N.i.K. and S.i.K./Maritime Southeast Asia/Mauritius (Fig. 9). Nevertheless, most of the shared major Mafa-A lineage groups were found to be linked to different major Mafa-B lineage groups when animals native to mainland N.i.K. and S.i.K./Maritime Southeast Asia/Mauritius were compared (Fig. 10). This example illustrates the dynamic diversity of the Mhc class I region in different populations of cynomolgus macaques. Nine of the major Mafa-A lineage groups were found to share linkage to a particular Mafa-B lineage group when animals native to mainland N.i.K. and S.i.K/Maritime Southeast Asia/Mauritius were compared (Fig. 10). For instance, major Mafa-A lineage group A1*006 shares linkage to B*002 and B*013. However, when we zoomed in on the haplotypes, and taking all the gene and allelic variation into account, we found that animals native to mainland N.i.K. and S.i.K. ultimately have their own characteristic Mafa-A and/or Mafa-B gene/allele combinations. The major Mafa-B lineage groups B*013, B*028, B*045, and B*056 are frequently observed and found in linkage to a variety of major Mafa-A lineage groups in animals native to mainland N.i.K. and S.i.K./Maritime Southeast Asia/Mauritius (Fig. 10). This might suggest that these B lineage groups have been enriched in the different populations and may encode molecules that are of benefit to the species.
Schematic presentation of evolutionary dynamics in Mafa-A/B haplotypes in cynomolgus macaque populations native to the mainland north of the isthmus of Kra (North) and mainland south of the isthmus of Kra /Maritime Southeast Asia/Mauritius (South). The 37 major Mafa-A lineage groups shared between north and south populations are taken as a starting point (Table S5) and are shown on the left-hand side of the figure. For all currently documented Mafa-A/B haplotypes (this study) (Budde et al. 2010; Karl et al. 2017; Otting et al. 2012; Saito et al. 2012; Shiina et al. 2015; Shortreed et al. 2020), the major Mafa-B lineage groups linked to a particular major Mafa-A lineage group are indicated, and each major B lineage group was given its specific background color. The major B lineage groups indicated with a white background color were found only in combination with one major A lineage group. The underscored major Mafa-A lineage groups share one or more linked major Mafa-B lineage group(s) between the north and south populations, and the Mafa-B lineage group(s) in question is/are indicated in boldface and outlined with a black border
Among the populations originating from mainland N.i.K. and S.i.K./Maritime Southeast Asia/Mauritius, and for which as well as the Mafa-A/B also the Mafa-DRB/DQ/DP haplotypes are characterized, we observed that two of the DRB haplotypes are shared (Fig. 11A) (this study) (Otting et al. 2012; Shortreed et al. 2020). Of these, Mafa-DRB1*03:03/DRB*W001:01/DRB*W003:02/DRB6*01:28 was frequently observed, eleven times in the population native to mainland N.i.K. and seven times in the population native to mainland S.i.K./Maritime Southeast Asia/Mauritius. For both populations, linkage to -DQA1*01:03:02 was often seen, whereas allelic variation was detected in the -DQB1 gene and lineage and allelic variation for the -DPA1 and -DPB1 genes (Fig. 11A). Furthermore, in most cases, this specific DRB-haplotype is found linked to different major Mafa-A and Mafa-B lineage groups. Only the combination with A1*043/B*045 is observed in both cohorts, north and south (Fig. 12), but as mentioned above, when we zoomed in on the haplotype, we found allelic variation for the Mafa-A and Mafa-B region configurations. For two animals originating from mainland N.i.K., we found that the DRB-haplotype is linked to the same major Mafa-A and Mafa-B lineage groups (Fig. 12; Table S1, haplotype nos. 5 and 6). However, these two animals differ with regard to the -DQB1, -DPA1, and -DPB1 genes (Fig. 11A, gene combinations indicated by a gray background) as well as with regard to their mitochondrial 12S rRNA gene segment. The other shared DRB-haplotype, DRB*W005:01/DRB*W021:01/DRB6*01:01, is found linked to different DQ and DP genes in the geographically distinct populations (Fig. 11A) and is linked to different major Mafa-A and Mafa-B lineage groups (Fig. 12). In addition, five DRB region configurations were found to be shared (Fig. 11B), eventually exhibiting their own specific allelic variation, depending on the animal/population of origin it was characterized in. Therefore, the majority of the DRB haplotypes in cynomolgus macaques of different geographic origins are characteristic for a population, which has its influences on the haplotype dynamics that can be observed between the cynomolgus macaque populations native to mainland N.i.K. and mainland S.i.K./Maritime Southeast Asia/Mauritius.
DRB haplotypes (A) and region configurations (B) shared between cynomolgus macaque populations of different geographical origin. A The column “DRB #” provides the number of Mafa-A/B/DRB/DQ/DP haplotypes on which the two indicated DRB haplotypes were characterized in a specific population. The -DQA/-DQB/-DPA/-DPB combinations found linked to the indicated DRB haplotype are shown, followed by the number (number sign) of times the combination has been found in a specific population studied. The gray background highlights the combination of genes that are found in two unrelated animals in combination with the indicated DRB-haplotype and the major Mafa-A1*003/B*001 lineage group combination (Fig. 12; Table S1, haplotype nos. 5 and 6)
Schematic presentation of the two DRB-haplotypes shared between cynomolgus macaque populations native to the mainland north of the isthmus of Kra (North) and mainland south of the isthmus of Kra/Maritime Southeast Asia/Mauritius (South) and the different major Mafa-A and Mafa-B lineage groups that can be found linked to it. Color codes for the different major Mafa-B lineage groups correspond to the colors in Fig. 10. Those major Mafa-A lineage groups indicated with a number sign and the major Mafa-B lineage groups shown in a white box are only documented in one of the populations. The underscored major Mafa-A/B lineage group combination is found in two unrelated animals together with the indicated DRB-haplotype (Table S1, haplotype nos. 5 and 6)
Discussion
The extensive characterization of the Mhc class I and II repertoire in different populations of cynomolgus macaques by a variety of research groups has resulted in data on 51 genes for which 2977 different alleles have been archived (IPD-MHC NHP release version 3.7.0.0). It is evident that particular Mhc class I and II alleles may be shared between cynomolgus macaque populations of different geographic origins (this study) (Karl et al. 2017; Otting et al. 2017) and even between different macaque species (Doxiadis et al. 2006; Karl et al. 2017). Despite this sharing of allelic entities, our present communication illustrates that the Mafa-A/B/DRB/DQ/DP haplotypes might be characteristic of a given cynomolgus macaque population. These observations suggest dynamic evolution of Mhc haplotypes in cynomolgus macaques of different geographic origins. Such novel haplotypes might be generated by the formation of population specific alleles; the generation of novel region configurations for A, B, or DRB by recombination events; recombination between different Mafa-A, Mafa-B, Mafa-DRB, Mafa-DQ, and Mafa-DP haplotypes; or a combination of these genetic events. All alleles shared between the two populations studied in this communication are, however, found embedded in different Mafa-A/B/DRB/DQ/DP haplotypes (Table S1).
To elucidate the geographic origin of the 84 newly introduced cynomolgus macaques into BPRC’s existing breeding colony, the mitochondrial 12S rRNA gene segment was analyzed. However, mitochondrial DNA is inherited strictly from the maternal line, and therefore, we want to emphasize that for defining the geographic origin of an animal; in addition, pedigree details might be required to link the ancestry of an MHC haplotype to a geographic origin.
The microsatellite analysis revealed four Mhc-A STR patterns being shared between the two cohorts that we investigated (Table S1). The subsequent sequencing analyses showed that only in the case of pattern 188/167, also an identical Mafa-A1 allele — namely, Mafa-A1*038:01:01 — was segregating in both cohorts (Table S1, haplotype nos. 42/43 and 90, respectively). This substantiates the notion that sequencing results involving newly analyzed populations/animals are initially required for a correct calling of a Mhc allele that is segregating with a particular STR pattern. If once mapped, however, STR typing is extremely cost-efficient to determine the Mhc variability inherited by offspring and can be applied for instance in colony management (de Groot et al. 2017a; Doxiadis et al. 2007; Otting et al. 2012).
Some haplotypes in cynomolgus macaques were found to contain two distinct copies of a Mafa-A1 gene, which may exhibit dissimilar transcription profiles as well. For example, we observed a dissimilar transcription profile for two closely related Mafa-A1*011 alleles. Mafa-A1*011:01 seems to represent a major (Table S1, haplotype nos. 22 and 23), whereas Mafa-A1*011:02 was found lowly transcribed on two different haplotypes in addition to the highly transcribed Mafa-A1*001:01:02 or Mafa-A2*24:07 (Table S1, haplotype nos. 1 and 79). These observations highlight the plasticity of the macaque A1 region. To determine what might possibly cause the aberrant transcription level, further research is required on those haplotypes characterized with an additional copy of an A1 gene.
Disparities in transcription levels are frequently observed in the Mhc class I B genes, and dissimilar transcription profiles for different alleles belonging to a certain B lineage have been found. An example are members of the Mafa-B*056 lineage. Mafa-B*056:01:01 and Mafa-B*056:07 are both found to be transcribed abundantly and are considered as “major 1” (Table S1, haplotype nos. 11, 45, 53, 61, 64, 73, 75, and 79 and haplotype no. 70, respectively), whereas Mafa-B*056:04 and Mafa-B*056:02:01 are embedded within haplotypes next to the highly transcribed Mafa-B*001:02 and Mafa-B*017:01 alleles, respectively (Table S1, haplotype no. 38 and haplotype nos. 82, 83, and 85, respectively). Moreover, within the present panel, we encountered examples of an allele that may display a disparate transcription profile, and this seems to be haplotype dependent. For instance, Mafa-B*023:03 is found in animals native to mainland N.i.K. as a “major 1” B allele, whereas in an animal native to mainland S.i.K./Maritime Southeast Asia, this allele is found as “major 2” on a haplotype linked to Mafa-B*017:01 as “major 1” B allele (Table S1, haplotype no. 18/69 and haplotype no. 82, respectively). Another example involves Mafa-B*018:01:01, which is considered as “major 1” on a haplotype characterized in an animal native to mainland S.i.K./Maritime Southeast Asia, whereas it is found as “major 3” on a haplotype next to Mafa-B*017:02:01/-B*007:01:01 (“major 1″/”major 2″) in animals native to mainland N.i.K. (Table S1, haplotype no. 84 and nos. 7, 39, 55, and 56, respectively). Therefore, identical alleles embedded within different haplotypes may show dissimilar transcription profiles. It is probable that recombination processes underlie this. As a result of these types of processes, one may hypothesize that an allele might be placed for instance in front of a stronger or weaker promoter, thereby causing the difference in transcription observed. It has long been known that different macaque Mhc class I alleles show disparate transcription levels, which have been substantiated by different studies showing that this also translates to the level of cell surface expression (de Groot et al. 2017b; Otting et al. 2005; Rosner et al. 2010; Wiseman et al. 2009). To date, however, nothing is known about the mechanisms that regulates this phenomenon.
The comparison of the Mafa-A/B haplotypes between animals native to mainland N.i.K. and mainland S.i.K./Maritime Southeast Asia/Mauritius suggests that certain major Mafa-A and Mafa-B lineage groups are only present in a population from a specific geographic region (Table S5). On the one hand, this might have been caused by the differential selection and different pathogenic pressures that these strictly separated populations may have experienced. On the other hand, we see the sharing of major Mafa-A and Mafa-B lineage groups (Fig. 10), and even the sharing of alleles between the two defined populations (Table S1), but always in the context of a different Mafa-A/B/DRB/DQ/DP haplotype, which might be considered a marker for a population. The different Mhc regions (Mafa-A, Mafa-B, Mafa-DRB, Mafa-DQA/DQB, and Mafa-DPA/DPB) of a Mafa-A/B/DRB/DQ/DP haplotype seem to be exchanged by recombination, resulting in the high number of cynomolgus macaque Mhc haplotypes that have been published, and with this number being expected to grow substantially.
The low transcription level observed for particular macaque A and B alleles can cause them to be missed in the method we currently use, which might result in haplotypes not being complete at this point. Furthermore, the current method lacks information on the exact order of the genes on a haplotype. Newly developed techniques, such as the target-enrichment of large genomic regions as well as whole genome sequencing using PacBio or Oxford Nanopore platforms, are now widely accessible. This might help and speed up our insight into the structure and number of genes present on an Mhc haplotype in the near future, as has been done recently for the KIR region (Bruijnesteijn et al. 2021). Moreover, these approaches give access to DNA modification profiles that could provide answers in the direction of the disparate transcription (expression) profile observed for certain macaque Mhc class I alleles. In conclusion, the present data strongly suggest that Mafa-A/B/DRB/DQ/DP haplotypes are characteristic of a population of animals from a specific geographic origin and may be a useful tool in conservation biology as well.
References
Aarnink A, Mee ET, Savy N, Congy-Jolivet N, Rose NJ, Blancher A (2014) Deleterious impact of feto-maternal MHC compatibility on the success of pregnancy in a macaque model. Immunogenetics 66:105–113
Almond N, Berry N, Stebbings R, Preston M, Ham C, Page M, Ferguson D, Rose N, Li B, Mee ET, Hassall M, Stahl-Hennig C, Athanasopoulos T, Papagatsias T, Herath S, Benlahrech A, Dickson G, Meiser A, Patterson S (2019) Vaccination of macaques with dna followed by adenoviral vectors encoding simian immunodeficiency virus (SIV) gag alone delays infection by repeated mucosal challenge with SIV. J Virol 93:e00606-00619
Blancher A, Aarnink A, Savy N, Takahata N (2012a) Use of cumulative Poisson probability distribution as an estimator of the recombination rate in an expanding population: example of the macaca fascicularis major histocompatibility complex. G3 (Bethesda) 2:123–130
Blancher A, Aarnink A, Tanaka K, Ota M, Inoko H, Yamanaka H, Nakagawa H, Apoil PA, Shiina T (2012b) Study of cynomolgus monkey (Macaca fascicularis) Mhc DRB gene polymorphism in four populations. Immunogenetics 64:605–614
Blancher A, Aarnink A, Yamada Y, Tanaka K, Yamanaka H, Shiina T (2014) Study of MHC class II region polymorphism in the Filipino cynomolgus macaque population. Immunogenetics 66:219–230
Bontrop RE, Otting N, de Groot NG, Doxiadis GG (1999) Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 167:339–350
Boszormenyi KP, Stammes MA, Fagrouch ZC, Kiemenyi-Kayere G, Niphuis H, Mortier D, van Driel N, Nieuwenhuis I, Vervenne RAW, Haaksma T, Ouwerling B, Adema D, Acar RF, Zuiderwijk-Sick E, Meijer L, Mooij P, Remarque EJ, Oostermeijer H, Koopman G, Hoste ACR, Sastre P, Haagmans BL, Bontrop RE, Langermans JAM, Bogers WM, Kondova I, Verschoor EJ, Verstrepen BE (2021) The post-acute phase of SARS-CoV-2 infection in two macaque species is associated with signs of ongoing virus replication and pathology in pulmonary and extrapulmonary tissues. Viruses 13:1673
Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, Watkins DI (1996) The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol 156:4656–4665
Breitbach ME, Newman CM, Dudley DM, Stewart LM, Aliota MT, Koenig MR, Shepherd PM, Yamamoto K, Crooks CM, Young G, Semler MR, Weiler AM, Barry GL, Heimsath H, Mohr EL, Eichkoff J, Newton W, Peterson E, Schultz-Darken N, Permar SR, Dean H, Capuano S, 3rd, Osorio JE, Friedrich TC, O’Connor DH (2019) Primary infection with dengue or Zika virus does not affect the severity of heterologous secondary infection in macaques. PLoS Pathog 15:e1007766
Briata P, Radka SF, Sartoris S, Lee JS (1989) Alternative splicing of HLA-DQB transcripts and secretion of HLA-DQ beta-chain proteins: allelic polymorphism in splicing and polyadenylylation sites. Proc Natl Acad Sci U S A 86:1003–1007
Bruijnesteijn J, van der Wiel M, de Groot NG, Bontrop RE (2021) Rapid characterization of complex killer cell immunoglobulin-like receptor (KIR) regions using Cas9 enrichment and nanopore sequencing. Front Immunol 12:722181
Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O’Connor DH (2010) Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780
Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL, Bimber BN, O’Connor SL, O’Connor DH (2009) Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 61:177–187
Creager HM, Becker EA, Sandman KK, Karl JA, Lank SM, Bimber BN, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2011) Characterization of full-length MHC class II sequences in Indonesian and Vietnamese cynomolgus macaques. Immunogenetics 63:611–618
de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE (2004) Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol 172:6152–6157
de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE (2008) Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics 60:737–748
de Groot N, Doxiadis GG, Otting N, de Vos-Rouweler AJ, Bontrop RE (2014) Differential recombination dynamics within the MHC of macaque species. Immunogenetics 66:535–544
de Groot N, Stanbury K, de Vos-Rouweler AJ, de Groot NG, Poirier N, Blancho G, de Luna C, Doxiadis GG, Bontrop RE (2017) A quick and robust MHC typing method for free-ranging and captive primate species. Immunogenetics 69:231–240
de Groot NG, Heijmans CMC, de Ru AH, Janssen GMC, Drijfhout JW, Otting N, Vangenot C, Doxiadis GGM, Koning F, van Veelen PA, Bontrop RE (2017) A specialist macaque MHC class I molecule with HLA-B*27-like peptide-binding characteristics. J Immunol 199:3679–3690
de Groot NG, Otting N, Maccari G, Robinson J, Hammond JA, Blancher A, Lafont BAP, Guethlein LA, Wroblewski EE, Marsh SGE, Shiina T, Walter L, Vigilant L, Parham P, O’Connor DH, Bontrop RE (2020) Nomenclature report 2019: major histocompatibility complex genes and alleles of Great and Small Ape and Old and New World monkey species. Immunogenetics 72:25–36
Dijkman K, Vervenne RAW, Sombroek CC, Boot C, Hofman SO, van Meijgaarden KE, Ottenhoff THM, Kocken CHM, Haanstra KG, Vierboom MPM, Verreck FAW (2019) Disparate tuberculosis disease development in macaque species is associated with innate immunity. Front Immunol 10:2479
Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE (2006) Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 58:259–268
Doxiadis GG, de Groot N, Claas FH, Doxiadis II, van Rood JJ, Bontrop RE (2007) A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc Natl Acad Sci U S A 104:8907–8912
Doxiadis GG, de Groot N, de Groot NG, Rotmans G, de Vos-Rouweler AJ, Bontrop RE (2010) Extensive DRB region diversity in cynomolgus macaques: recombination as a driving force. Immunogenetics 62:137–147
Doxiadis GG, de Groot N, Otting N, Blokhuis JH, Bontrop RE (2011) Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics 63:73–83
Doxiadis GG, de Vos-Rouweler AJ, de Groot N, Otting N, Bontrop RE (2012) DR haplotype diversity of the cynomolgus macaque as defined by its transcriptome. Immunogenetics 64:31–37
Emborg ME (2017) Nonhuman primate models of neurodegenerative disorders. ILAR J 58:190–201
Glazko GV, Nei M (2003) Estimation of divergence times for major lineages of primate species. Mol Biol Evol 20:424–434
Huang S, Huang X, Li S, Zhu M, Zhuo M (2019) MHC class I allele diversity in cynomolgus macaques of Vietnamese origin. PeerJ 7:e7941
Karl JA, Graham ME, Wiseman RW, Heimbruch KE, Gieger SM, Doxiadis GG, Bontrop RE, O’Connor DH (2017) Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics 69:211–229
Kawamoto Y, Kawamoto S, Matsubayashi K, Nozawa K, Watanabe T, Stanley MA, Perwitasari-Farajallah D (2008) Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: an assessment of nuclear and mitochondrial DNA polymorphisms. J Med Primatol 37:45–54
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A 86:6196–6200
Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH (2005) Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239
Kwon Y, Lee KW, Park H, Son JK, Lee J, Hong J, Park JB, Kim SJ (2019) Comparative study of human and cynomolgus T-cell depletion with rabbit anti-thymocyte globulin (rATG) treatment-for dose adjustment in a non-human primate kidney transplantation model. Am J Transl Res 11:6422–6432
Li W, Wang T, Ling F, Zhao H, Wei L, Zhuo M, Du H, Wang X (2012) Identification of MhcMafa-DRB alleles in a cohort of cynomolgus macaques of Vietnamese origin. Am J Primatol 74:958–966
Ling F, Wei LQ, Wang T, Wang HB, Zhuo M, Du HL, Wang JF, Wang XN (2011) Characterization of the major histocompatibility complex class II DOB, DPB1, and DQB1 alleles in cynomolgus macaques of Vietnamese origin. Immunogenetics 63:155–166
Ling F, Zhuo M, Ni C, Zhang GQ, Wang T, Li W, Wei LQ, Du HL, Wang JF, Wang XN (2012) Comprehensive identification of high-frequency and co-occurring Mafa-B, Mafa-DQB1, and Mafa-DRB alleles in cynomolgus macaques of Vietnamese origin. Hum Immunol 73:547–553
Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, Ho CS, de Groot NG, Flicek P, Bontrop RE, Hammond JA, Marsh SG (2017) IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45:D860–D864
O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH (2007) Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59:449–462
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A 102:1626–1631
Otting N, de Groot N, de Vos-Rouweler AJ, Louwerse A, Doxiadis GG, Bontrop RE (2012) Multilocus definition of MHC haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 64:755–765
Otting N, van der Wiel MK, de Groot N, de Vos-Rouweler AJ, de Groot NG, Doxiadis GG, Wiseman RW, O’Connor DH, Bontrop RE (2017) The orthologs of HLA-DQ and -DP genes display abundant levels of variability in macaque species. Immunogenetics 69:87–99
Pendley CJ, Becker EA, Karl JA, Blasky AJ, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2008) MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 60:339–351
Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O’Brien SJ, Pecon-Slattery J (2011) A molecular phylogeny of living primates. PLoS Genet 7:e1001342
Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE (2020) IPD-IMGT/HLA Database. Nucleic Acids Res 48:D948–D955
Rosner C, Kruse PH, Lubke T, Walter L (2010) Rhesus macaque MHC class I molecules show differential subcellular localizations. Immunogenetics 62:149–158
Saito Y, Naruse TK, Akari H, Matano T, Kimura A (2012) Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 64:131–141
Sano K, Shiina T, Kohara S, Yanagiya K, Hosomichi K, Shimizu S, Anzai T, Watanabe A, Ogasawara K, Torii R, Kulski JK, Inoko H (2006) Novel cynomolgus macaque MHC-DPB1 polymorphisms in three South-East Asian populations. Tissue Antigens 67:297–306
Shiina T, Yamada Y, Aarnink A, Suzuki S, Masuya A, Ito S, Ido D, Yamanaka H, Iwatani C, Tsuchiya H, Ishigaki H, Itoh Y, Ogasawara K, Kulski JK, Blancher A (2015) Discovery of novel MHC-class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and Sanger sequencing: Mafa-class I polymorphism. Immunogenetics 67:563–578
Shortreed CG, Wiseman RW, Karl JA, Bussan HE, Baker DA, Prall TM, Haj AK, Moreno GK, Penedo MCT, O’Connor DH (2020) Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques. Immunogenetics 72:225–239
Sussman RW, Tattersall I (1986) Distribution, abundance, and putative ecological strategy of Macaca fascicularis on the Island of Mauritius, Southwestern Indian Ocean. Folia Primatol 46:28–43
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035
Tosi AJ, Morales JC, Melnick DJ (2003) Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57:1419–1435
Tosi AJ, Coke CS (2007) Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol Phylogenet Evol 42:498–504
van der Wiel MK, Otting N, Zeijdel LM, Doxiadis GG, Bontrop RE (2015) Novel DRA alleles extracted from seven macaque cohorts. Tissue Antigens 85:146–148
van der Wiel MKH, Doxiadis GGM, de Groot N, Otting N, de Groot NG, Poirier N, Blancho G, Bontrop RE (2018) MHC class I diversity of olive baboons (Papio anubis) unravelled by next-generation sequencing. Immunogenetics 70:439–448
Wang HB, Ling F, Zhuo M, Wang JF, Wang XN (2011) Twenty-three novel major histocompatibility complex class I B alleles identified in cynomolgus macaques of Vietnamese origin. Tissue Antigens 77:346–348
Westbrook CJ, Karl JA, Wiseman RW, Mate S, Koroleva G, Garcia K, Sanchez-Lockhart M, O’Connor DH, Palacios G (2015) No assembly required: full-length MHC class I allele discovery by PacBio circular consensus sequencing. Hum Immunol 76:891–896
Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E Jr, Wright C, Harkins T, O’Connor DH (2009) Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15:1322–1326
Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O’Connor DH (2013) Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J 54:196–210
Zhang GQ, Ni C, Ling F, Qiu W, Wang HB, Xiao Y, Guo XJ, Huang JY, Du HL, Wang JF, Zhao ShJ, Zhuo M, Wang XN (2012) Characterization of the major histocompatibility complex class I A alleles in cynomolgus macaques of Vietnamese origin. Tissue Antigens 80:494–501
Acknowledgements
We thank G. Doxiadis for constructive feedback, D. Devine for editing the manuscript, and F. van Hassel for preparing the figures.
Funding
This work was supported in part by the NIH/NIAID contract numbers HHSN272201100013C and HHSN272201600007C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Groot, N.G., de Groot, N., de Vos-Rouweler, A.J.M. et al. Dynamic evolution of Mhc haplotypes in cynomolgus macaques of different geographic origins. Immunogenetics 74, 409–429 (2022). https://doi.org/10.1007/s00251-021-01249-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01249-y