Abstract
Cynomolgus macaques (Macaca fascicularis) are used widely in biomedical research, and the genetics of their MHC (Mhc-Mafa) has become the focus of considerable attention in recent years. The cohort of Indonesian pedigreed macaques that we present here was typed for Mafa-A, -B, and -DR, by sequencing, as described in earlier studies. Additionally, the DRB region of these animals was characterised by microsatellite analyses. In this study, full-length sequencing of Mafa-DPA/B and -DQA/B in these animals was performed. A total of 75 different alleles were observed; 22 of which have not previously been reported, plus 18 extended exon 2 alleles that were already known. Furthermore, two microsatellites, D6S2854 and D6S2859, were used to characterise the complex Mafa-A region. Sequencing and segregation analyses revealed that the length patterns of these microsatellites are unique for each Mafa-A haplotype. In this work, we present a pedigreed colony of approximately 120 cynomolgus macaques; all of which are typed for the most significant polymorphic MHC class I and class II markers. Offspring of these pedigreed animals are easily characterised for their MHC by microsatellite analyses on the Mafa-A and -DRB regions, which makes the cumbersome sequencing analyses redundant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nonhuman primates are often used in biomedical research as animal models for a variety of human diseases. Traditionally, the rhesus macaque (Macaca mulatta) has been the preferred species. In recent years however, cynomolgus macaques (Macaca fascicularis), also known as long-tailed or crab-eating macaques, have emerged in a number of studies. Whereas the rhesus macaques originate in Southern Asia and China, the cynomolgus macaques are found in Southeast Asia, including the Filipinos, and on the East African Island of Mauritius. They are used in research on infectious diseases such as AIDS, tuberculosis, and dengue (Cafaro et al. 2010; Chen et al. 2007; Langermans et al. 2001; Mee et al. 2009). Moreover, they are applied as models for Alzheimer’s and Parkinson’s disease, and in transplantation studies (Emborg 2007; Haustein et al. 2010; Wang et al. 2007).
Because of the animals’ role as models for immunological disorders, the Major histocompatibility complex (MHC) of the cynomolgus macaque (MhcMafa) is the subject of investigations. The highly polymorphic genes of the MHC encode cell-surface proteins, of which the most relevant are the classical class I and class II molecules. These proteins play a critical role in immunological responses to pathogens and hence may have a profound impact on disease susceptibility and organ transplantation (Bontrop 2006; Parham and Ohta 1996). In addition, the genetics of the killer cell Ig-like receptor (KIR) polymorphisms are currently being studied in these animals (Bimber et al. 2008).
Molecular genetic analyses of the class I genes in different populations of cynomolgus monkeys have led to hundreds of full-length Mafa-A and -B alleles (Aarnink et al. 2011; Budde et al. 2010; Campbell et al. 2009; Kita et al. 2009; Krebs et al. 2005; Lawrence et al. 2012; Ling et al. 2012; Mitchell et al. 2011; Otting et al. 2007, 2009; Pendley et al. 2008; Saito et al. 2011; Uda et al. 2004; Wiseman et al. 2009; Zhuo et al. 2011); all of which have been assembled in the IPD-MHC database (de Groot et al. 2012; Robinson et al. 2003). These studies have shown that most alleles are specific for the geographic origins of the animals, a phenomenon that is also observed in rhesus macaques (Otting et al. 2005, 2008). This observation dictates that each cohort of animals that is used in biomedical research be investigated for its repertoire of MHC alleles. A relatively restricted MHC diversity is present in macaques of Mauritian origin, which makes them useful in studies that require MHC-identical animals (Budde et al. 2010; Cafaro et al. 2010; Krebs et al. 2005; Mee et al. 2009). The class I alleles observed in this homogenous population are shared with those of animals originating in Indonesia, which confirms the founder effect as regards animals that originated in this archipelago.
The most intensively investigated class II genes are DRA and -B, DQA and -B, and DPA and -B (Bontrop et al. 1999; Creager et al. 2011; de Groot et al. 2004, 2008; Doxiadis et al. 2007, 2006; Hashiba et al. 1993; Leuchte et al. 2004; Ling et al. 2011; O’Connor et al. 2007; Otting et al. 2002; Sano et al. 2006). The proteins are expressed as heterodimers, consisting of alpha and beta chains that are encoded by the A and B genes. In recent years, sequencing analyses of class II genes have been performed mostly on exon 2, since these polymorphic exons encode the peptide binding domains of the heterodimers. However, the alignments of full-length class II sequences in the IPD-MHC database reveal alleles that share exon 2 but that differ in other exons (Creager et al. 2011). In comparison to class I alleles in the cynomolgus macaques, the class II alleles appear to be less specific for the geographic origins of the animals.
The Biomedical Primate Research Centre (BPRC) houses a population of cynomolgus macaques that has been pedigreed for up to seven generations. Most animals were sequenced for the class I A and B (Otting et al. 2007, 2009) and for the class II DR regions (Doxiadis et al. 2006), and these analyses were supplemented with microsatellite analyses for the DRB region (de Groot et al. 2008; Doxiadis et al. 2010). From a subset of these animals, DQB sequencing of exon 2 has been performed (Doxiadis et al. 2006; Otting et al. 2002). With the aim of completing the map of haplotypes, we performed sequencing analyses of DPA, DPB, DQA, and DQB genes. Since polymorphism of class II genes may be present in exons other than exon 2, we performed full-length sequencing. Furthermore, with the intention of avoiding future time-consuming sequencing analyses on newborn animals in this colony, the Mafa-A region was characterised by microsatellite analyses (Doxiadis et al. 2011a; Penedo et al. 2005).
Materials and methods
Animals and cell lines
The BPRC houses a self-sustaining colony of about 120 cynomolgus macaques. Before being transferred to the BPRC facilities, the animals were pedigreed in 12 matrilines for up to seven generations at the University of Utrecht. Beginning with ten animals in the late 1960s, the colony was gradually extended by a few males and three females in the 1970s, nine females and their offspring in the 1980s, and three males in the 1990s, all from different institutions. The estimated number of founder animals is therefore at least 30. The animals originated in the Indonesian islands and continental Malaysia, as defined by mitochondrial (12S RNA) analyses (Doxiadis et al. 2006).
Lymphoblastoid B-cell-lines and genomic DNA are available from most animals in the colony. In addition, samples are present from related animals that are no longer available. The animals were typed earlier for Mafa-A, -B, and -DR (Doxiadis et al. 2011b; Otting et al. 2007, 2009), and the combination of these data resulted in 30 different haplotypes. Since mitochondrial DNA and MHC haplotypes are inherited independently and the founder animals of the colony have died, it is not possible to distinguish unambiguously between Indonesian and Malaysian MHC haplotypes. For these reasons, all haplotypes are considered as Indonesian. To sequence the full-length DP and DQ sequences, we chose at least three animals, preferably from different matrilines, for each haplotype in the colony. The genomic DNA samples of all animals were included for Mafa-A microsatellite or short tandem repeat (STR) analyses.
cDNA, cloning, and sequencing
RNA was isolated from B-lymphocytes (Rneasy kit, Qiagen) and subjected to One-Step RT-PCR, as recommended by the supplier (Promega). The primers used for the amplification of DPA, DPB, DQA, and DQB transcripts were copied from a study on Mauritian cynomolgus macaques (O’Connor et al. 2007). Additional primers are DPA-F-inex: GACAGAATGTTCSAGACCAG, DPA-R-inex: CGTTGTCTCAGGSATCTGGAT, DPB-F-inex: GGCRTTACTGATGGTRCTGC, DPB-R-inex: CCTCCTGTGCATGAAGATGCCC, DQA-F-inex: CAAAGCTCTGWTGCTGGSGG, and DQA-R-inex: GCCCTTGGTGTCTGGARGC. The final elongation step was extended to 7 min to generate a 3′dA overhang. The RT-PCR products were cloned using the PCR cloning kit (Qiagen). After transformation, 16 colonies were selected for plasmid isolation. Sequencing reactions were performed using the BigDye terminator cycle sequencing kit, and samples were run on an automated capillary sequencing system (Applied Biosystems Genetic Analyzer 3130).
Sequences were analyzed using MacVector™ version 10.6.0 (Oxford Molecular Group), followed by manual adjustments. New alleles, represented by three identical clones, present in at least two different animals or RT-PCR samples, were submitted to the EMBL-EBI and, for official designations, to the nonhuman primate section of the IMGT/MHC Immuno Polymorphism Database (de Groot et al. 2012; Robinson et al. 2003).
Mafa-A microsatellite typing
The polymerase chain reaction (PCR) amplifications, with primers for the microsatellites D6S2854, a (TAAA)n repeat, and D6S2859, a mixed dinucleotide (TA)×(CA)y repeat, were performed under the same conditions as described previously for the rhesus macaques at BPRC (Doxiadis et al. 2011a). Briefly, both PCR reactions on genomic DNA samples were multiplexed in a 25-μl mixture. The amplified DNA fragments were mixed with size standards and separated on an automated capillary electrophoresis system (Applied Biosystems Genetic Analyzer 3130). Fragment-length analysis was performed using the Genemapper software (Applied Biosystems).
Results
Novel alleles
Mafa-DPA1/DPB1
In the panel of cynomolgus macaques, 17 different Mafa-DPA1 sequences were detected. All DPA1 alleles were amplified with the MHCII-DPA-F/R primer pair (O’Connor et al. 2007). For animals that showed only one allele in the RT-PCR, an extra amplification was performed with the DPA-inex (in exon) primers. These inex-primers were designed so that they anneal to the outermost exons of all known alleles in the IPD-MHC database. In some instances, the second allele was found, indicating that certain alleles may be amplified abundantly, at the expense of the one present on the other haplotype. These minor alleles were sufficiently present in other animals, however. Eight alleles had not been reported previously, whereas another one extended a known DPA1 exon 2 sequence. These alleles were submitted to the EMBL/EBI and, for official designations, to the IPD-MHC database. The sequences are listed in Table 1, including the accession numbers, reference animals, and the geographic origin of the animals.
The DPB1 amplification was initiated with the MHCII-DPB-F1/R1 primer pair (O’Connor et al. 2007), though for those samples with only one allele, we performed second RT-PCRs with another reverse primer: MHCII-DPB-R2. In the event that this did not yield a second allele, the samples were subjected to RT-PCR with DPB-inex primers that anneal to the exons in all known Mafa-DPB sequences. Using these primers, four alleles were found, which were not detected with the full-length primers in any of the animals. A total of 21 Mafa-DPB1 sequences were observed within the cohort (Table 1). Four alleles were submitted as novel sequences, whereas three others extended known exon 2 sequences. Two of the latter are not full-length class II sequences because they were detected using the DPB-inex primers.
Mafa-DQA/DQB
Twenty Mafa-DQA alleles were present in the cohort, of which nine were novel, and five extended known exon 2 sequences. The analyses were initiated using the DQA-F2/R2 primers and extended with DQA-F1/R1 and DQA-inex primers in the event that alleles were not amplified. Finally, 17 DQB alleles were found with DQB-F2/R2; nine of which extended known exon 2 sequences. One allele was submitted as a novel sequence.
Among the 75 sequences detected, 16 are shared with alleles of Mauritian origin (O’Connor et al. 2007). This is not surprising, since the founders of this colony are assumed to have originated in the Indonesian archipelago. Seven alleles were also detected in animals of Vietnamese descent. This confirms earlier findings of shared class II alleles in animals of Vietnamese and Indonesian descent (Creager et al. 2011). One allele was seen earlier in a Malaysian cohort (Aarnink et al. 2011).
Mafa-A microsatellite typing
The microsatellites D6S2854 and D6S2859 have been used earlier to type the different groups of rhesus macaques housed at BPRC. These analyses have shown that both markers are polymorphic in length and that the typing results are reproducible. Genotyping resulted in patterns of varying length that, based on segregation analyses, are associated with the Mamu-A haplotypes. Since all of these haplotypes and associated microsatellite patterns are known within the rhesus breeding groups, this technique is applied as a fast typing method for screening the animals’ offspring. The cynomolgus and rhesus macaques share the organisation of the MHC class I region (Otting et al. 2007; Watanabe et al. 2007), and the same microsatellite method was expected to be suitable for genotyping the cohort of cynomolgus macaques. Indeed, the length patterns of the D6S2854 and D6S2859 PCR fragments matched with the 30 Mafa-A/-B/-DRB haplotypes in the cohort.
MhcMafa haplotypes
Based on earlier studies on the class I A and B as well as on the class II DR regions, it was possible to define 30 haplotypes within the colony of animals. In this study, we were able to append the DP and DQ full-length alleles in addition to the D6S2854 and D6S2859 microsatellite length patterns. An overview of the haplotypes is provided in Table 2. The gene regions are listed according to their position on the chromosome, which is as follows: Mafa-A, -I and -B, -DRA, -DRB, -DQA1, -DQB1, -DPA1, and DPB1, respectively (Watanabe et al. 2007). The exact order of loci within the duplicated gene regions, such as Mafa-B and -DRB, is unknown; hence, the listing of these alleles in the columns is arbitrary. The haplotypes are ordered vertically based on the Mafa-A1 alleles and are numbered on an arbitrary basis. The list of 30 is extended by two more haplotypes that appear to be combinations of others. Number 4 is composed of haplotypes 3 and 21, whereas 22 may be the result of a crossing between 21 and 26. These recombined haplotypes are observed in four and two animals, respectively.
The Mafa-A, -B, and -I alleles were detected by full-length sequencing on RNA, and thus, they represent only transcribed genes (Otting et al. 2009). The Mafa-A, -B, and -I alleles are unique for each haplotype, with a few exceptions. The combinations Mafa-A1*031:01/A2*05:04 and Mafa-B*033:03/B*090:01 are both observed in two haplotypes. Furthermore, two other alleles Mafa-B*104:04 and Mafa-B*147:01 are seen in two haplotypes, however, in combination with different other Mafa-B alleles. Two Mafa-A4*14 sequences, indicated by the lineage-number only, are not confirmed by three identical clones, and the exact full-length sequences are still unknown.
All haplotypes may contain an allele of the oligomorphic Mafa-I gene, a locus that was first described in the rhesus macaque (Urvater et al. 2000). However, in most cases, the criterion of three identical full-length clones to define an allele was not fulfilled. As mentioned previously, each Mafa-A/-B/-I combination is associated with a unique microsatellite length pattern. The sizes of the D6S2854 and D6S2859 repeats are depicted in base pairs in the columns next to the Mafa-A1 columns in Table 2. The lengths are based on comparison to size standards in the capillary electrophoresis and may differ slightly from the actual length of the PCR products. However, the position of the repeats within the Mafa-A region, in relation to the different loci Mafa-A1, -A2, and so on, is unknown. In 2010, the Mafa-B sequences that were submitted to the Nonhuman Primate part of the IPD-MHC database underwent a drastic renaming, with the aim of adjusting the lineage numbers to Mamu-B alleles in the rhesus macaques. Hence, the Mafa-B allele-names in Table 2 are different from those listed in the original paper that was published in 2009 (Otting et al. 2009). The old and new designations are available at http://www.ebi.ac.uk/ipd/mhc/nhp/nomenclature.html.
In contrast to the class I sequences, the sharing of alleles among haplotypes is observed within the DR region. The DRA*01:02:01:01 allele, for example, is present in 10 haplotypes, whereas *01:09 and *01:02:21 are unique for haplotypes 30 and 24, respectively. The DRA alleles are detected by full-length sequencing on RNA (Doxiadis et al. 2011b). In contrast to the DRA locus, of which only one copy is present per haplotype, the DRB region in macaques is well known for its wide variety of region configurations (de Groot et al. 2004, 2008). Region configurations are groups of haplotypes that differ in the number and contents of DRB loci. Within this cohort of animals, the DRB region has been investigated thoroughly, by sequencing of exon 2 on genomic DNA as well as by full-length sequencing on RNA (Doxiadis et al. 2010, 2011b). The alleles that are transcribed, as pointed out by full-length sequencing on RNA, are depicted in boldface in Table 2. The number of DRB loci varies from 2 to 5, as is evident in the haplotypes 1 and 15, respectively. Five DRB combinations are shared by haplotypes, with one example present in 3, 9, and 32, and another in 8, 11, and 17. However, these shared DRB combinations may show differences in DRA alleles as well as in microsatellite lengths. The microsatellite D6S2878, used for DRB-typing, is located in intron 2 of DRB genes. In contrast to the STRs in the Mafa-A region, it was possible to link each DRB allele to a particular STR length, by sequencing exon 2/intron 2 combinations. For these reasons, each column with DRB alleles in Table 2 has an adjacent column with the associated STR lengths.
The DP and DQ sequences that were generated in this study are listed in the last columns of Table 2. Several animals were subjected to repeated analyses, but for unknown reasons, we failed to detect DPA and DQB alleles corresponding to haplotype 10. Furthermore, haplotype 32 is based on one animal only, in which the other chromosome contained haplotype 9. Sequencing analyses in this animal yielded only DPA1*04:01/DPB1*02:05. It is possible that this animal is homozygous for the DP region, but it cannot be excluded that the second alleles were missed due to primer inconsistencies. An investigation of related animals is not possible, since this animal has no offspring in the BPRC population. In contrast to the class I sequences, the detected DQ and DP alleles are not always haplotype specific. The combination DPA1*02:03/DPB1*04:01, for example, is observed in three haplotypes, whereas DQA1*23:01/DQB1*18:04 is present in four combinations (combined haplotypes excluded). Other combinations, such as DPA1*02:20/DPB1*15:05 and DQA1*01:02/DQB1*06:07:01, are haplotype specific. When DQ and DP alleles are taken together, however, each haplotype appears to contain a unique combination of DQA/DQB/DPA/DPB alleles.
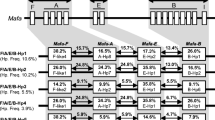
As previously mentioned, the BPRC cohort is a subset of a colony that had been housed and pedigreed at the University of Utrecht for up to seven generations. In this cohort, 121 animals (242 haplotypes) are fully typed for MHC class I and class II markers. However, genomic DNA samples and lymphoblastoid B cells, as well as pedigree data pertaining to the original colony, were available. Hence, it was possible to extend the number of typed animals to 195 (390 haplotypes). In the last columns of Table 2, the haplotype distributions are presented, in absolute numbers. The percentages are depicted in Fig. 1. Comparison of these percentages shows that the cohort has not experienced a dramatic shift in the haplotype distribution. A reduction is observed for haplotype 28, with a percentage of 4.4 % in the original group and 1.7 % in the BPRC cohort. The other differences are all negligible. Three haplotypes, 22, 29, and 32, with low frequencies in the original population, are not present in the current cohort. Two animals are homozygous for their MHC; one of them has haplotype 3, the most common in the cohort, whereas the other is homozygous for haplotype 21.
Concluding remarks
Here, we present cynomolgus macaques that are completely sequenced for the most significant MHC class I and class II genes, resulting in 32 distinct haplotypes. The estimated number of founding animals is around 30, which makes the number of haplotypes high in comparison to other cohorts (Mitchell et al. 2011). Additionally, the Mafa-A and -DRB regions are characterised by microsatellite typing. Using these high-throughput microsatellite techniques makes it easy to analyze the offspring of animals with these Mafa-A and -DRB markers and hence to deduce the accessory haplotype. Crossover events between Mafa-A and -DRB will be easily detected by using these techniques, and in these cases, further sequencing analyses of the particular infant can be performed. These techniques have already led to the definition of the two haplotypes 4 and 22, which may have been the result of recombination in recent generations. Similar observations were made in cynomolgus macaques of Mauritian origin (O’Connor et al. 2007). Crossover events on the centromeric side of the DRB region, however, are not detected with current microsatellite typing. Additional sequencing of DPB may be necessary to exclude these events, together with the investigation of polymorphic microsatellites on the centromeric side of the MHC region. The STR’s D6S2876, DS62747, D6S2745, and D6S2771, used by Mitchell and coworkers, may be appropriate candidates to type the class II region. These analyses may also be a starting point for comparison of our cohort to the Indonesian animals investigated by this research group (Mitchell et al. 2011).
This MHC-typed cohort of animals facilitates the choice of appropriate animals for research on immune-related diseases. Haplotypes 19 and 20, for example, are identical to the Mauritian haplotypes M1 and M3 (Budde et al. 2010), also known as H1 and H3 (Mee et al. 2009). These haplotypes are relevant to SIV pathogenesis and vaccine evaluation and are present in 12 and 4 individuals in the BPRC cohort, respectively.
References
Aarnink A, Apoil PA, Takahashi I, Osada N, Blancher A (2011) Characterization of MHC class I transcripts of a Malaysian cynomolgus macaque by high-throughput pyrosequencing and EST libraries. Immunogenetics 63:703–713
Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O’Connor DH (2008) Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol 181:6301–6308
Bontrop RE (2006) Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum Immunol 67:388–397
Bontrop RE, Otting N, de Groot NG, Doxiadis GG (1999) Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 167:339–350
Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O’Connor DH (2010) Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780
Cafaro A, Bellino S, Titti F, Maggiorella MT, Sernicola L, Wiseman RW, Venzon D, Karl JA, O’Connor D, Monini P, Robert-Guroff M, Ensoli B (2010) Impact of viral dose and major histocompatibility complex class IB haplotype on viral outcome in Mauritian cynomolgus monkeys vaccinated with Tat upon challenge with simian/human immunodeficiency virus SHIV89.6P. J Virol 84:8953–8958
Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL, Bimber BN, O’Connor SL, O’Connor DH (2009) Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 61:177–187
Chen L, Ewing D, Subramanian H, Block K, Rayner J, Alterson KD, Sedegah M, Hayes C, Porter K, Raviprakash K (2007) A heterologous DNA prime-Venezuelan equine encephalitis virus replicon particle boost dengue vaccine regimen affords complete protection from virus challenge in cynomolgus macaques. J Virol 81:11634–11639
Creager HM, Becker EA, Sandman KK, Karl JA, Lank SM, Bimber BN, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2011) Characterization of full-length MHC class II sequences in Indonesian and Vietnamese cynomolgus macaques. Immunogenetics 63:611–618
de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE (2004) Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol 172:6152–6157
de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE (2008) Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics 60:737–748
de Groot N, Otting N, Robinson J, Blancher A, Lafont BAP, Marsh SG, O’Connor D, Shiina T, Walter L, Watkins DI, Bontrop R (2012) Nomenclature report on the major histocompatibility complexes, genes, and alleles of Great Ape, Old and New World monkey species. Immunogenetics. doi:10.1007/s00251-012-0617-1
Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE (2006) Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 58:259–268
Doxiadis GG, de Groot N, Claas FH, Doxiadis II, van Rood JJ, Bontrop RE (2007) A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc Natl Acad Sci U S A 104:8907–8912
Doxiadis GG, de Groot N, de Groot NG, Rotmans G, de Vos-Rouweler AJ, Bontrop RE (2010) Extensive DRB region diversity in cynomolgus macaques: recombination as a driving force. Immunogenetics 62:137–147
Doxiadis GG, de Groot N, Otting N, Blokhuis JH, Bontrop RE (2011a) Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics 63:73–83
Doxiadis GG, de Vos-Rouweler AJ, de Groot N, Otting N, Bontrop RE (2011b) DR haplotype diversity of the cynomolgus macaque as defined by its transcriptome. Immunogenetics 64:31–37
Emborg ME (2007) Nonhuman primate models of Parkinson’s disease. ILAR J 48:339–355
Hashiba K, Kuwata S, Tokunaga K, Juji T, Noguchi A (1993) Sequence analysis of DPB1-like genes in cynomolgus monkeys (Macaca fascicularis). Immunogenetics 38:462
Haustein S, Kwun J, Fechner J, Kayaoglu A, Faure JP, Roenneburg D, Torrealba J, Knechtle SJ (2010) Interleukin-15 receptor blockade in non-human primate kidney transplantation. Transplantation 89:937–944
Kita YF, Hosomichi K, Kohara S, Itoh Y, Ogasawara K, Tsuchiya H, Torii R, Inoko H, Blancher A, Kulski JK, Shiina T (2009) MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaque. Immunogenetics 61:635–648
Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH (2005) Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239
Langermans JA, Andersen P, van Soolingen D, Vervenne RA, Frost PA, van der Laan T, van Pinxteren LA, van den Hombergh J, Kroon S, Peekel I, Florquin S, Thomas AW (2001) Divergent effect of bacillus Calmette-Guerin (BCG) vaccination on Mycobacterium tuberculosis infection in highly related macaque species: implications for primate models in tuberculosis vaccine research. Proc Natl Acad Sci U S A 98:11497–11502
Lawrence J, Orysiuk D, Prashar T, Pilon R, Fournier J, Rud E, Sandstrom P, Plummer FA, Luo M (2012) Identification of 23 novel MHC class I alleles in cynomolgus macaques of Philippine and Philippine/Mauritius origins. Tissue Antigens 79:306–307
Leuchte N, Berry N, Kohler B, Almond N, LeGrand R, Thorstensson R, Titti F, Sauermann U (2004) MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens 63:529–537
Ling F, Wei LQ, Wang T, Wang HB, Zhuo M, Du HL, Wang JF, Wang XN (2011) Characterization of the major histocompatibility complex class II DOB, DPB1, and DQB1 alleles in cynomolgus macaques of Vietnamese origin. Immunogenetics 63:155–166
Ling F, Zhuo M, Ni C, Zhang GQ, Wang T, Wei LQ, Du HL, Wang JF, Wang XN (2012) Comprehensive identification of high-frequency and co-occurring Mafa-B, Mafa-DQB1, and Mafa-DRB alleles in cynomolgus macaques of Vietnamese origin. Hum Immunol 73:547–553
Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, Verschoor EJ, Heeney JL, Le Grand R, Titti F, Almond N, Rose NJ (2009) Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 61:327–339
Mitchell JL, Mee ET, Almond NM, Cutler K, Rose NJ (2011) Characterisation of MHC haplotypes in a breeding colony of Indonesian cynomolgus macaques reveals a high level of diversity. Immunogenetics 64:123–129
O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH (2007) Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59:449–462
Otting N, de Groot NG, Doxiadis GG, Bontrop RE (2002) Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics 54:230–239
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A 102:1626–1631
Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE (2007) MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 59:367–375
Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG, Bontrop RE (2008) A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics 60:507–514
Otting N, Doxiadis GG, Bontrop RE (2009) Definition of Mafa-A and -B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 61:745–753
Parham P, Ohta T (1996) Population biology of antigen presentation by MHC class I molecules. Science 272:67–74
Pendley CJ, Becker EA, Karl JA, Blasky AJ, Wiseman RW, Hughes AL, O’Connor SL, O’Connor DH (2008) MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 60:339–351
Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, de Groot N, de Groot NG, Ward T, Doxiadis GG (2005) Microsatellite typing of the rhesus macaque MHC region. Immunogenetics 57:198–209
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31:311–314
Saito Y, Naruse TK, Akari H, Matano T, Kimura A (2011) Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 64:131–141
Sano K, Shiina T, Kohara S, Yanagiya K, Hosomichi K, Shimizu S, Anzai T, Watanabe A, Ogasawara K, Torii R, Kulski JK, Inoko H (2006) Novel cynomolgus macaque MHC-DPB1 polymorphisms in three South-East Asian populations. Tissue Antigens 67:297–306
Uda A, Tanabayashi K, Yamada YK, Akari H, Lee YJ, Mukai R, Terao K, Yamada A (2004) Detection of 14 alleles derived from the MHC class I A locus in cynomolgus monkeys. Immunogenetics 56:155–163
Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarczyk MS, Golos TG, Hughes AL, Bontrop RE, Watkins DI (2000) Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol 164:1386–1398
Wang CY, Finstad CL, Walfield AM, Sia C, Sokoll KK, Chang TY, Fang XD, Hung CH, Hutter-Paier B, Windisch M (2007) Site-specific UBITh amyloid-beta vaccine for immunotherapy of Alzheimer’s disease. Vaccine 25:3041–3052
Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, Kimura T, Soeda E, Torii R, Ogasawara K, Kulski JK, Inoko H (2007) A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 89:402–412
Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E Jr, Wright C, Harkins T, O’Connor DH (2009) Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15:1322–1326
Zhuo M, Wang HB, Ling F, Wang JF, Wang XN (2011) Eighteen novel MHC class I A alleles identified in Vietnamese-origin cynomolgus macaques. Tissue Antigens 78:139–142
Acknowledgments
The authors would like to thank Ms. Donna Devine for editing the manuscript. This study was supported in part by the NIH/NIAID contract numbers HHSN266200400088C/NOI-AI-0088 and 5R24RR016038-05 (CFA:03.9389).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Otting, N., de Groot, N., de Vos-Rouweler, A.J.M. et al. Multilocus definition of MHC haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 64, 755–765 (2012). https://doi.org/10.1007/s00251-012-0632-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-012-0632-2