Abstract
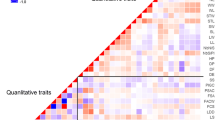
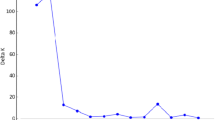
The distinctness, uniformity and stability (DUS) requirements involve expensive, space- and time-consuming measurements of morphological traits. Moreover, for a majority of traits, interactions between genotype and environment complicate the evaluation. Molecular markers have a potential to facilitate this procedure, increase the reliability of decisions, and substantially save the time and space needed for experiments. We chose 25 varieties of pea (Pisum sativum L.) from the list of recommended varieties for cultivation in the Czech Republic, and made both a standard classification by 12 morphological descriptors and a classification by biochemical-molecular markers. Two isozyme systems, 10 microsatellite loci, 2 retrotransposons for multilocus inter-retrotransposon amplified polymorphism (IRAP), and 12 retrotransposon-based insertion polymorphism (RBIP) DNA markers were analysed. The main objective of the study was to examine the potential of each method for discrimination between pea varieties. The results demonstrate a high potential and resolving power of DNA-based methods. Superior in terms of high information content and discrimination power were SSR markers, owing to high allelic variation, which was the only biochemical-molecular method allowing clear identification of all varieties. Retrotransposon markers in RBIP format proved to be the most robust and easy to score method, while multilocus IRAP produced informative fingerprint already in a single analysis. Isozyme analysis offered a fast and less expensive alternative. The results showed that molecular identification could be used to assess distinctness and complement morphological assessment, especially in cases where the time frame plays an important role. Currently developed pea marker systems might serve also for germplasm management and genetic diversity studies.
Similar content being viewed by others
References
Bredemeijer GMM, Cooke RJ, Ganal MW, Peeter R, Isakk P, Noordijk Y, et al. 2002. Construction and testing of a microsatellite database containing more than 500 tomato varieties. Theor Appl Genet 105: 1019–1026.
Burstin J, Charcosset A, 1997. Relationship between phenotypic and marker distances: theoretical and experimental investigation. Heredity 79: 477–483.
Dillmann C, Bar-Hen A, Guerin D, Charcosset A, Murigneux A, 1997. Comparison of RFLP and morphological distances between maizeZea mays L. inbred lines. Theor Appl Genet 95: 92–102.
Doyle JJ, Doyle JL, 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15.
Ford R, Le Roux K, Itman C, Brouwer JB, Taylor PWJ, 2002. Diversity analysis and genotyping inPisum with sequence-tagged microsatellite site (STMS) primers. Euphytica 124: 397–405.
Jing RC, Knox MR, Lee JM, Vershinin AV, Ambrose M, Ellis THN, Flavell AJ, 2005. Insertional polymorphism and antiquity of PDR1 retrotransposon insertions inPisum species. Genetics 171: 741–752.
Kwon YS, Lee J, Yi GB, Yi SI, Kim KM, Soh EH, et al. 2005. Use of SSR markers to complement tests of distinctiveness, uniformity and stability (DUS) of pepper (Capsicum annuum L.) varieties. Mol Cells 19: 428–435.
Law JR, Donini P, Koebner RMD, Reeves JC, Cooke RJ, 1998. DNA profiling and plant variety registration. III: The assessment of distinctness in wheat using amplified fragment length polymorphisms. Euphytica 102: 335–342.
Lefebvre V, Goffinet B, Chauvet JC, Caromel B, Signoret P, Brand R, Palloix A, 2001. Evaluation of genetic distances between pepper inbred lines for cultivar protection purposes: comparison of AFLP, RAPD and phenotypic data. Theor Appl Genet 102: 741–750.
Lombard V, Dubreuil P, Dillman C, Baril C, 2001. Genetic distance estimators based on molecular data for plant registration and protection: A review. Acta Hort 546: 55–63.
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, et al. 2005. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111: 1022–1031.
Mantel N, 1967. The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
Nuel G, Baril C, Robin S, 2001. Varietal distinctness assisted by molecular markers: A methodological approach. Acta Hort 546: 65–71.
Nybom H, 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecology 13: 1143–1155.
Pavelková A, Moravec J, Hájek D, Bareš I, Sehnalová J, 1986. Descriptor list of the genusPisum L. RICP Prague — Ruzyně, Genové zdroje 32: 46.
Posvec Z, Griga M, 2000. Utilization of isozyme polymorphism for cultivar identification of 45 commercial peas (Pisum sativum L.). Euphytica 113: 251–258.
Reif JC, Melchiger AE, Frish M, 2005. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci 45: 1–7.
Rohlf FJ, 2005. Numerical taxonomy and multivariate analysis system (NTSYS-pc) version 2.2. User guide. Exeter Software, New York, USA: 31.
Roldán-Ruíz I, van Eeuwijk FA, Gilliland TJ, Dubreuil P, Dillmann C, Lallemand J, et al. 2001. A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor Appl Genet 103: 1138–1150.
Saunders JA, Mischke S, Leamy EA, Hemeida AA, 2004. Selection of international molecular standards for DNA fingerprinting ofTheobroma cacao. Theor Appl Genet 110: 41–47.
Smýkal P, 2006. Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J Appl Genetic 47: 221–230.
Smýkal P, Valledor L, Rodriguez R, Griga M, 2007. Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Rep 26: 1985–1998.
Święcicki WK, Wolko B, 1987. Application of electro-phoretic methods of isozymes separation to genetical characterization of pea (Pisum sativum L. s. lat.) cultivars. Genet Pol 28: 89–99.
Święcicki WK, Wolko B, Apisitwanich S, Krajewski P, 2000. An analysis of isozymic loci polymorphism in the core collection of the PolishPisum genebank. Genet Res Crop Evol 47: 583–589.
Tessier C, David J, This P, Boursiquot JM, Charrier A, 1999. Optimization of the choice of molecular markers for varietal identification inVitis vinifera L. Theor Appl Genet 98: 171–177.
Tommasini L, Batley J, Arnold GM, Cooke RJ, Donini P, Lee D, et al. 2003. The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106: 1091–1101.
[UPOV] International Union for the Protection of New Varieties of Plants, 1990. Guidelines for the Conduct of Tests for Distinctness, Uniformity, and Stability. Document UPOV TG/4/7. Geneva, Switzerland.
[UPOV] 1991. International convention for the protection of new varieties of plants, Publication No. 221e, March 19, Geneva, Switzerland.
[UPOV] BMT/36/10,2002. Progress report of the 36th session of the Technical Committee, the Technical Working Parties and Working Group on biochemical and molecular techniques and DNA-profiling in particular. Geneva, Switzerland.
van Eeuwijk FA, Baril CP, 2001. Conceptual and statistical issues related to the use of molecular markers for distinctness and essential derivation. Acta Hort 546: 35–53.
van Treuren R, van Hintum TJL, 2001. Identification of intra-accession genetic diversity in selfing crops using AFLP markers: implications for collection management. Genet Res Crop Evol 48: 287–295.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Smýkal, P., Horáček, J., Dostálová, R. et al. Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J Appl Genet 49, 155–166 (2008). https://doi.org/10.1007/BF03195609
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03195609