Abstract
A set of 59 spring barley introgression lines (ILs) was developed from the advanced backcross population S42. The ILs were generated by three rounds of backcrossing, two to four subsequent selfings, and, in parallel, marker-assisted selection. Each line includes a single marker-defined chromosomal segment of the wild barley accession ISR42-8 (Hordeum vulgare ssp. spontaneum), whereas the remaining part of the genome is derived from the elite barley cultivar Scarlett (H. vulgare ssp. vulgare). Based on a map containing 98 SSR markers, the IL set covers so far 86.6% (1041.5 cM) of the donor genome. Each single line contains an average exotic introgression of 39.2 cM, representing 3.2% of the exotic genome. The utility of the developed IL set is illustrated by verification of QTLs controlling resistance to powdery mildew (Blumeria graminis f. sp. hordei L.) and leaf rust (Puccinia hordei L.) which were previously identified in the advanced backcross population S42. Altogether 57.1 and 75.0% of QTLs conferring resistance to powdery mildew and leaf rust, respectively, were verified by ILs. The strongest favorable effects were mapped to regions 1H, 0–85 cM and 4H, 125–170 cM, where susceptibility to powdery mildew and leaf rust was decreased by 66.1 and 34.7%, respectively, compared to the recurrent parent. In addition, three and one new QTLs were localized, respectively. A co-localization of two favorable QTLs was identified for line S42IL-138, which holds an introgressed segment in region 7H, 166–181. Here, a reduction effect was revealed for powdery mildew as well as for leaf rust severity. This line might be a valuable resource for transferring new resistance alleles into elite cultivars. In future, we aim to cover the complete exotic genome by selecting additional ILs. We intend to conduct further phenotype studies with the IL set in regard to the trait complexes agronomic performance, malting quality, biotic stress, and abiotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cultivated barley (Hordeum vulgare ssp. vulgare, hereafter abbreviated with Hv) is one of the four most important cereals worldwide and is mainly used as feed grain and for malt production. The domestication of barley took place approximately 10,000 years ago in the Fertile Crescent from its wild relative Hordeum vulgare ssp. spontaneum (hereafter abbreviated with Hsp, Badr et al. 2000). As illustrated by Tanksley and McCouch (1997), the domestication of crops has led to a dramatic loss of allelic diversity. As a result, exotic alleles from related wild species become more and more valuable for breeding of improved crop varieties, in regard to qualitative as well as quantitative traits like disease resistances and yield-related traits, respectively. The development of introgression lines (ILs) provides a promising opportunity to efficiently use the genetic potential of wild species. As described by Zamir (2001), a complete set of introgression lines is supposed to represent the entirety of the exotic genome, while each line contains only a single marker-defined chromosomal segment of the exotic parent and the remaining genome is consistently derived from the elite parent. First IL sets in plants were developed for tomato (Eshed et al. 1992; Eshed and Zamir 1994; Fulton et al. 1997) and rice (Jena et al. 1992; Aida et al. 1997). In recent years, complete or nearly complete IL sets were developed for further tomato (Monforte and Tanksley 2000; Canady et al. 2005; Finkers et al. 2007) and rice wild species (Li et al. 2005; Tian et al. 2006a; Tan et al. 2007), as well as Arabidopsis thaliana (Keurentjes et al. 2007), the D-genome of wheat (Pestsova et al. 2006), maize (Szalma et al. 2007), and melon (Eduardo et al. 2005). Also in other crop species, lines containing introgressed segments from wild species (e.g., near-isogenic lines, recombinant, or backcross inbred lines) were generated and used for genetic studies. But here, the coverage of the donor genome was not complete and/or the lines contained multiple exotic introgressions simultaneously. Examples for these lines can be found for instance for cabbage (Ramsay et al. 1996), coffee (Prakash et al. 2002), cotton (Percy et al. 2006), lettuce (Jeuken and Lindhout 2004), peanut (Garcia et al. 1995), pepper (Ben Chaim et al. 2003), rapeseed (Howell et al. 1996), and soybean (Concibido et al. 2003). For spring barley, Brown et al. (1988) primarily generated a set of 84 backcross lines, each carrying a single homozygous isozyme marker-defined segment of an Hsp accession. Matus et al. (2003) developed 140 recombinant chromosome substitution lines (RCSLs) for spring barley by introgression of Hsp alleles. The lines, selected on the basis of genotype data from 47 SSR markers, contained on average 12.6% Hsp genome, and mostly carried several Hsp introgressions simultaneously. A similar population was generated by Hori et al. (2005). Here, altogether 134 RCSLs with an averaged proportion of Hsp genome of 12.9% were genotyped with 25 SSR and 60 EST markers and subsequently applied to QTL analysis. A core-set of 19 RCSLs represented the complete Hsp genome, where each line included one or several exotic introgressions. Valuable plant material for the development of a complete IL set in barley was also generated by von Korff et al. (2004). Two sets of candidate introgression lines (pre-ILs) were selected from two BC2DH populations, using the Hsp accession ‘ISR42-8’ as donor and the spring barley cultivars ‘Scarlett’ (S) and ‘Thuringia’ (T) as recurrent parents. The selection of 49 (S42) and 43 (T42) pre-ILs was carried out on the basis of genotype data from 98 SSRs. The sets of selected lines carried 98.1 and 93.0% of the exotic genome, and contained on average 3.0 and 2.5 introgressions (S42 and T42, respectively).
The numerous advantages of ILs are described in detail by Zamir (2001). Due to the very small portion of exotic genome, epistatic and linkage drag effects are reduced, and the phenotypic variation between the ILs can be attributed with high accuracy to the particular introduced segment. Furthermore, once a complete homozygous IL set is developed, it is a reliable and stable genetic resource, and each IL can be used directly for breeding. To date, ILs are applied to a multitude of genetic studies. The Solanum pennellii IL population, established by Eshed and Zamir (1994), is so far the best characterized. Here, almost 3,000 putative QTLs for different trait complexes like yield-associated traits, (Eshed and Zamir 1995; Hanson et al. 2007); morphological traits (Holtan and Hake 2003; Semel et al. 2006), and metabolites (Baxter et al. 2005; Overy et al. 2005; Schauer et al. 2006) were identified. For yield-related traits, several QTL studies were also conducted in rice (e.g. Mei et al. 2006; Tan et al. 2007) and wheat (Liu et al. 2006; Pestsova et al. 2006). As it was reported by several publications, ILs were also applied to the analysis of quality parameters like amount of antioxidants in tomato fruits (Rousseaux et al. 2005); milling quality of rice grains (Zheng et al. 2007), amount of high molecular weight glutenin subunits in wheat grains (Liu et al. 2007) and post-harvest decay and quality of melon fruits (Eduardo et al. 2007; Fernandez-Trujillo et al. 2007; Obando et al. 2008). ILs were also tested under abiotic stress conditions to detect QTLs for drought tolerance (Xu et al. 2005; Zhang et al. 2006; Zhou et al. 2006; Siangliw et al. 2007), and tolerance to salinity and phosphorus deficiency in rice (Li et al. 2005). Not only quantitative traits were dissected by ILs, but also qualitative traits, which are affected by a single or a few loci. Examples for investigation of disease resistances are described for wheat (Leonova et al. 2007; Simón et al. 2007; Song et al. 2007), but also for tomato (Finkers et al. 2007). The above-mentioned barley S42 pre-ILs were evaluated with regard to disease resistances, agronomic performance, and malting quality (von Korff et al. 2005, 2006, 2008).
In the present paper, we report on the first development of an IL set for barley. Each IL carries a single introgression of the exotic Hsp accession ‘ISR42-8’ in the genetic background of the elite spring barley cultivar ‘Scarlett’. The set is generated by backcrossing, selfing, and marker-assisted selection. In order to illustrate the applicability of the spring barley ILs, the lines were used for verification of QTLs for field resistance against powdery mildew (Blumeria graminis f. sp. hordei L.) and leaf rust (Puccinia hordei L.).
Materials and methods
Development and molecular characterization of introgression lines
The project started with 40 pre-ILs (candidate introgression lines), which were selected from 301 BC2DH lines of the spring barley population S42 (von Korff et al. 2004). The lines were generated from a primary cross between the spring barley cultivar ‘Scarlett’ (Hordeum vulgare ssp. vulgare) and the wild barley accession ISR42-8 from Israel (Hordeum vulgare ssp. spontaneum).
For the development of the final introgression lines, the selected 40 pre-ILs were backcrossed once again with ‘Scarlett’ (BC3) in order to further reduce the portion of the Hsp genome and to minimize the target introgression (Fig. 1). The BC3 plants were subsequently selfed twice to achieve recombined homozygous lines. In BC3S2, all plants were genotyped with a total of 98 SSR markers as described by von Korff et al. (2004). On average, 140 plants per line were investigated with, on average, 12 informative SSRs, which revealed the Hsp genotype in the appropriate pre-IL. Based on these genotype data, useful plants were selected as introgression lines. The criteria for selection were as follows: (1) a line contains a single continuous Hsp introgression and carried the Hv allele at all remaining SSR loci. (2) Within the IL set, the introgressions should overlap to ensure that the complete exotic genome is represented. All finished ILs were verified again with SSR markers in BC3S4. Here, the two outer loci of the Hsp introgression and one random locus, which revealed the exotic allele in the corresponding pre-IL, were investigated again to confirm the homozygous exotic genotype of each line. The ILs were subsequently propagated until BC3S6 to obtain a sufficient number of seeds for phenotype studies in 2007. In this generation, the ILs were verified again on the basis of 30 random SSR loci, which were not investigated before, to confirm the pure genetic Hv background. The size of the Hsp introgression was calculated for each finished IL. Here, the half-intervals flanking a marker locus were considered to be of the same genotype. For calculation of the portion of exotic germplasm per IL, a total genome size of 1,202 cM was assumed on the basis of the applied SSR map (von Korff et al. 2004).
Strategy for development and evaluation of introgression lines from population S42. After several rounds of backcrossing and selfing, a set of introgression lines, originating from the cross ‘Scarlett’ × ‘ISR42-8’, was selected by marker-assisted selection (MAS). The genotypes of the ILs were verified in subsequent generations. Afterward, the lines were evaluated with regard to disease resistances. Finally, the genotype and phenotype data of the ILs were used for line × phenotype association studies to localize QTL effects and favorable Hsp introgressions
Evaluation of powdery mildew and leaf rust
In order to evaluate the selected S42ILs in regard to their resistance to powdery mildew and leaf rust, field tests with 39 lines were conducted at two different locations in Germany in 2007. The locations were the experimental research station Dikopshof of the University of Bonn (D07, West Germany), and the breeder’s experimental field station Gudow (G07, Nordsaat Saatzucht, North Germany). The ILs were cultivated in three replications of two rows per line. As a control, the recurrent parent ‘Scarlett’ was tested in 12 replications of two rows. In addition, the spring barley cultivar ‘Barke’ was tested in nine replications to compare the performance of the S42ILs with a current elite cultivar. The field management was in accordance with local practice, but without applying fungicides. No artificial infection with pathogens was carried out. The severity of disease symptoms was surveyed on leaves at the maximum stage of disease development on a scale from one (no symptoms) to nine (completely covered with symptoms).
Genetic correlations
Pearson’s correlation coefficient (r) was calculated with the CORR procedure, implemented in the software package SAS Enterprise Guide 4.1 (SAS Institute 2006). The genetic correlations between leaf rust and powdery mildew were calculated based on least squares means (LSMEANS) for each of the 39 S42ILs averaged across replications and environments.
Line × phenotype association study
The data for powdery mildew and leaf rust were analyzed with SAS Enterprise Guide 4.1 (SAS Institute 2006). For detection of QTLs and identification of favorable Hsp introgressions, a two-factorial mixed model analysis of variance (ANOVA) was carried out with the general linear model (GLM) procedure. The GLM model is stated below:
where μ is the general mean, L i is the fixed effect of the ith line, E j is the random effect of the jth environment, L × E ij is the random interaction effect of the ith line and the jth environment and ε k(ij) is the error term of the phenotype Y ijk , calculated from k replications. When the ANOVA revealed significant differences with P < 0.05 between lines or line × environment interactions, a Dunnett multiple comparison test of LSMEANS differences between the ILs was conducted with the recurrent parent ‘Scarlett’ as the control (Dunnett 1955). A QTL main effect for powdery mildew or leaf rust resistance was accepted, when the disease score of a particular IL was significantly different from ‘Scarlett’ across all environments with P < 0.05. A QTL × environment interaction effect was accepted, when the disease score of a particular IL was significantly different from ‘Scarlett’ in at least one of the environments with P < 0.05. If several ILs with overlapping or flanking introgressions showed significant effects of the same direction, a single QTL was assumed to cause the effect. Overlapping introgressions contained at least one common SSR locus, whereas flanking introgressions covered adjacent loci. The relative performance of an introgression line [RP(IL)] was calculated as follows: RP(IL) = [LSMEANS(IL) − LSMEANS(Scarlett)] × 100/LSMEANS(Scarlett). For each line the LSMEANS were calculated across replications and environments as stated above.
Results
Selection and characterization of introgression lines
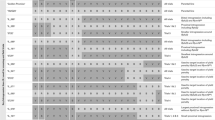
As a result of the marker-assisted selection in BC3S2 and BC3S4, a set of 59 S42ILs was developed (Table 1). The ILs carry single exotic introgressions from the Hsp accession ‘ISR42-8’. Four of these lines, S42IL-102, -114, -123, and S42IL-129, revealed a second, non-target, introgression on chromosomes 2H, 5H, 4H, and 1H, respectively (Fig. 2). Furthermore, two target introgressions on chromosome arm 3HL and 4HS were each represented by two lines, S42IL-114 and S42IL-140 and S42IL-118 and S42IL-120, respectively. Both pairs remain in the final IL set since it is expected that future high-resolution genotyping will reveal differences in introgression size between the independent sister lines.
Graphical genotypes of 59 Hsp introgression lines from population S42. The horizontal lines represent the seven barley chromosomes with 98 SSR loci. The order of SSRs is in agreement with von Korff et al. (2004). The 59 S42ILs, the recurrent parent ‘Scarlett’ (Hv) and the donor parent ‘ISR 42-8’ (Hsp) are illustrated vertically. The order of the S42ILs is in agreement with Table 1. Loci, which carry the homozygous Hsp genotype are depicted in grey, and heterozygous loci are illustrated in black. The remaining genome is derived from the recurrent parent ‘Scarlett’ (indicated in white)
Based on the SSR map published by von Korff et al. (2004), 1,041.5 cM (86.6% of 1,202 cM) of the exotic Hsp genome is covered by S42ILs. A similar estimate can be found by counting exotic SSR alleles. Altogether 87 (88.8%) Hsp alleles at the 98 SSR loci are represented by at least one IL, whereas 11 exotic SSR alleles are not yet covered by S42ILs. Chromosome 6H of the exotic parent is completely represented by ten different S42ILs. Chromosomes 4H and 7H are largely covered by ILs (92.1 and 85.9% genome coverage per chromosome, respectively). The selected S42ILs contain on average exotic introgressions of 39.2 cM (range 9.0–134.0 cM), representing 3.2% of the exotic genome (range 0.7–11.1%). Thirteen lines carry the Hsp allele only at a single SSR locus, whereas in 27 ILs the exotic segment includes more than three linked marker loci. In all 59 selected ILs, the introgression covers on average 3.4 SSR loci. Twenty-five S42ILs are unique. They represent the minimum number of S42ILs to cover the Hsp genome. The 34 non-unique lines are partial duplicates with shorter introgressions, which are useful for fine-mapping of markers and QTLs. Examples are S42ILs -106, -107 and -108, possessing introgressions on the top of chromosome 2H. S42IL-108 represents a unique introgression, extending from 17 to 92 cM. In contrast, the non-unique introgressions of the remaining two ILs extend from 17 to 27 cM and from 17 to 42 cM, respectively, and are thus already represented in the first IL.
Field experiment for powdery mildew and leaf rust resistance
In season 2007, 39 S42ILs were tested for reactions to powdery mildew and leaf rust infection in the field at two different locations. Across both environments, the mean score of powdery mildew symptoms per S42IL ranged from 1.7 to 7.0, whereas ‘Scarlett’ exhibited a mean disease score of 4.9 (Table 2). The check ‘Barke’ showed a mean disease score of 1.1. For leaf rust, the average disease severity for the S42ILs ranged from 2.7 to 5.7 scores across both environments. For ‘Scarlett’, a mean value of 4.1 scores was recorded, and the check ‘Barke’ exhibited a mean disease score of 2.8. The field experiment revealed only a weak correlation (r = 0.25 with P = 0.02) between scores for powdery mildew and leaf rust diseases based on LSMEANS for 39 S42ILs calculated across two environments and three replications.
Identification and verification of QTLs for powdery mildew resistance
The 39 S42ILs were subjected to a line × phenotype association study in order to verify QTL effects, which were previously detected with the advanced backcross population S42, as well as to identify new QTLs. The mixed model ANOVA for powdery mildew resistance revealed significant line effects (P < 0.001) and line × environment interaction effects (P = 0.003, data not shown). Ten introgression lines revealed powdery mildew scores significantly different from the control ‘Scarlett’. As illustrated in Fig. 3, the Hsp introgressions of these lines are located on chromosomes 1H, 2H, 4H, and 7H. Taking into account that some introgressions overlapped or were flanked, a total of seven QTLs for powdery mildew resistance could be assumed. Two QTLs are presumably located on chromosome 1H since the S42ILs-101 and -104 do not overlap. One QTL, represented by S42IL-110, is located on chromosome 2H. Three further QTLs are located on chromosome 4H. Although the two corresponding S42ILs -123 and -124 do overlap, their effects are contrasting (Table 3). Finally, at least one QTL is located on chromosome 7H. Although the two introgressions present in S42ILs -135 and -138 do not share a common SSR allele, their Hsp introgressions might potentially overlap in the border interval 155–166 cM. Six QTLs could be detected both as line effect and as L × E interaction effect (Table 3), whereas one QTL, present in S42IL-118, was only detected as line L × E interaction effect. The strongest favorable effect was measured for QPm.S42IL-1H.a, which was identified by S42ILs -101 and -102. Here, the Hsp introgression located on chromosome 1H, 0–14 cM and 1H, 0–85 cM, respectively, was associated with a reduction of disease severity by 52.5% (2.6 scores) and 66.1% (3.3 scores), respectively, relative to the control ‘Scarlett’. For both lines, the QTL effect was highly significant (P < 0.001) across both environments and also in each single environment (Table 2). As shown in Table 3, QPm.S42IL-1H.a verified the favorable QTL effect QPm.S42-1H.a, which was detected in the S42 population by von Korff et al. (2005). It was located in the chromosomal region 1H, 0–28 cM and caused a reduced powdery mildew susceptibility of 51.5%. One additional favorable QTL effect was localized in S42ILs as well as in the S42 population on chromosome 7H (QPm.S42IL-7H.a and QPm.S42-7H.a, respectively). Here, the disease severity was reduced by 22.0 and 31.9%, respectively, in comparison to ‘Scarlett’. The three lines S42IL-110, S42IL-118, and S42IL-123, which carry Hsp introgressions on chromosome 2H, 80–86 cM; 4H, 31–57 cM; and 4H, 125–170 cM, respectively, revealed a significant higher susceptibility against powdery mildew in comparison to ‘Scarlett’. As illustrated in Table 3, the disease severity was increased by 25.4% (1.3 scores), 28.6% (1.3 scores) and 42.4% (2.1 scores), respectively. Two of these effects (QPm.S42IL-2H.a and QPm.S42IL-4H.b) correspond to QPm.S42-2H.b and QPm.S42-4H.a, which were mapped by von Korff et al. (2005) to the regions 2H, 42–86 cM and 4H, 125–132 cM. They revealed an unfavorable Hsp effect, increasing powdery mildew symptoms by 17.0 and 16.1%, respectively.
SSR map showing the Hsp introgressions of 39 S42ILs and illustrating significant line × phenotype associations for resistance to powdery mildew and leaf rust. Chromosomes are shown as black bars with cM values for SSR loci following the order of von Korff et al. (2004). The extent of Hsp introgressions are given in grey bars right to the chromosomes. Significant associations are illustrated as symbols below the S42ILs. They either exhibit a reduction (filled symbols) or increase (empty symbols) in disease symptoms caused by the Hsp introgression. Associations solely detected as line × environment interaction effects are highlighted by an asterisk right to the symbol. S42ILs -110, -123 and -138 on chromosomes 2H, 4H and 7H, respectively, revealed two QTL effects each
Identification and verification of QTLs for leaf rust resistance
The mixed model ANOVA for leaf rust resistance revealed significant line effects (P < 0.001), but no significant line × environment interaction effect (P = 0.1855, data not shown). Altogether, four QTL effects, present in five S42ILs, were detected for leaf rust resistance (Table 3; Fig. 3). Two QTLs were located on chromosome 2H and one QTL each on chromosome 4H and 7H. Three QTLs were simultaneously detected as line effect and L × E interaction effect. One QTL was solely detected as L × E interaction effect. The strongest favorable effect of an Hsp introgression was measured in S42IL-138 (QLr.S42IL-7H.a). Here, the disease severity was reduced in environment D07 by 41.5% (1.4 scores). As illustrated in Table 3, this QTL effect confirmed QLr.S42-7H.a, which was identified in the S42 population by von Korff et al. (2005). Here, a significant marker main effect was detected in region 7H, 166–181. The disease symptoms were reduced by 28.9%, relative to ‘Scarlett’. The overlapping S42ILs -122 and -123 with Hsp introgressions located on chromosome 4H, 125–132 cM and 4H, 125–170 cM, respectively, revealed reductions in disease severity by 34.7% (-1.4 scores) each. In population S42, QLr.S42-4H.a was also mapped to the region 4H, 125–132 and showed a significant reduction of leaf rust symptoms by 37.6% (von Korff et al. 2005). The strongest unfavorable Hsp effect was measured in S42IL-107 on chromosome 2H, 17–42 cM (QLr.S42IL-2H.b). In this region, an increase of 38.8% (1.6 scores) in leaf rust susceptibility was detected. S42IL-110, which also carries an introgression on 2H from 80–86 cM, showed an increase in leaf rust symptoms by 30.6% (1.3 scores) in comparison to ‘Scarlett’. This effect corresponds to QLr.S42-2H.a, which was detected in the S42 population, and showed a significantly reduced susceptibility of −10.1% at the locus 2H, 86 cM (von Korff et al. 2005).
Discussion
Selection and characterization of introgression lines
In the present study, a set of 59 introgression lines of the population S42 was generated by three rounds of backcrossing, each theoretically reducing the portion of the exotic genome by 50% in the followed generation. Two or four selfings were subsequently performed to derive completely homozygous lines, which are a stable genetic resource for further evaluation. As described by von Korff et al. (2004), a pre-selection step, based on genotype data of 98 SSR markers, was carried out in the BC2DH generation. Here, a set of 40 candidate introgression lines were selected from a population consisting of 301 lines. The set presented the complete Hsp genome by a minimum number of lines, each containing a portion of the exotic genome as small as possible. Pure S42ILs were finally identified after a second selection step in the BC3S2 or BC3S4 generation. One or several pre-selection steps are generally advisable to reduce the required population size. Therefore, the propagation and genotyping of a large number of unusable recombinant plants can be avoided. A pre-selection in previous backcross generations, either based on genotype or phenotype data, has also proved to be helpful for the selection of IL sets in other crop species. For instance, Eshed and Zamir (1994) conducted two phenotypic selection steps in BC1 to BC1S5 generation based on horticultural characteristics, followed by two rounds of genotypic selection in BC1S6 to BC4S1 generation. For melon, Eduardo et al. (2005) pursued a selection strategy similar to those of the S42ILs. They initially performed a selection of 25 double haploid lines, based on the proportion of the donor genome. Several selection steps in subsequent backcross generations were followed, and finally a total of 57 near-isogenic lines (NILs) were predominantly selected in the BC3S1 or BC3S2 generation. In general, most IL sets were finally selected after up to ten generations, including three or four rounds of backcrossing (e.g. Szalma et al. 2007; Tian et al. 2006a).
So far, 86.6% (1,041.5 cM) of the Hsp genome is covered by pure S42ILs. Introgression lines, covering the missing chromosomal regions, will be selected in future within BC3 or BC4 progenies. For this purpose, a pool of 45 potential ILs, which are still under selection, is available. The selection of more ILs with smaller exotic introgressions is also advisable, since there are several S42ILs which possess very large introgressions. Therefore, all pure S42ILs will be backcrossed again with ‘Scarlett’, and appropriate recombinants will be identified and enhanced as so called Sub-ILs. Subsequently, selected Sub-ILs containing very small introgressions and carrying interesting QTLs, will be directly used for fine-mapping and, eventually, map-based cloning. Several reports for different crop species underline the usefulness of ILs for these purposes. By means of ILs, yield-associated QTLs were fine-mapped in tomato (Eshed and Zamir 1995), rice (Tian et al. 2006b), and wheat (Röder et al. 2008). Two studies in tomato focused on the dissection of regions containing QTLs for fruit quality and shape (Yates et al. 2004; van der Knaap et al. 2004, respectively). Furthermore, the tomato fruit color gene Beta, the fruit weight QTL fw2.2, and the sugar yield QTL Brix9-2-5 were cloned based on IL mapping and fine-mapping (Ronen et al. 2000; Frary et al. 2000; Fridman et al. 2004, respectively).
In future, the genotyping of the S42ILs with Illumina SNP chip markers (Rostoks et al. 2006) or Diversity Arrays Technology (DArT) markers (Wenzl et al. 2006) will foster the fine-mapping of the introgressions. So far, the average genomic resolution of the S42IL set is 12.5 cM/SSR. The least marker density of 21.4 cM/SSR was detected for the long arm of chromosome 6H. An increased genomic resolution is also necessary to confirm that the S42ILs do not possess additional non-target introgressions. Despite these prospective demands, the described set of 59 S42ILs is comparable to IL sets developed for other crop species, with regard to the ‘quality’ of introgression lines, e.g., exotic genome coverage and proportion of exotic genome present in each line. Among so far generated IL sets, the IL library of Solanum pennellii in S. esculentum, is the furthest developed set. Eshed and Zamir (1994) selected 50 ILs, which cover the whole genome of the exotic species S. pennellii. Based on the analysis with 350 RFLP markers, all ILs were verified as pure lines, each carrying single exotic segments of 33 cM in average. Currently, the S. pennellii IL set consists of 76 different lines covering the complete tomato genome (Lippman et al. 2007). In Arabidopsis thaliana, a set of 92 near-isogenic lines (NILs) containing one to four introgressions of the Cape Verde Islands (Cvi) accession within the genetic background of Landsberg erecta (Ler), was developed by Keurentjes et al. (2007). The target introgressions included on average 31.7 cM. Twenty-five lines, selected as a core-set, covered >90% of the donor genome. Eduardo et al. (2005) reported on the development of 57 NILs for melon, which altogether represented 85% of the introgressed exotic genome. The single lines exhibited an average introgression size of 41 cM and an average proportion of exotic genome of 3.4%. Also the genomic resolution of 15.7 cM/marker, provided by a map with 62 SSR markers, was similar to our results for the S42ILs. In comparison to IL sets of several other crop species, the S42ILs so far exhibited a high grade of pureness since 55 lines possess single exotic introgressions. These lines allow a precise localization of the chromosomal region, when the exotic introgression causes a phenotypic effect relative to the recurrent parent ‘Scarlett’.
Identification and verification of QTLs for powdery mildew and leaf rust resistance
As a first example for applications, the S42ILs were evaluated with regard to powdery mildew and leaf rust resistance. The aims were (1) to verify QTLs, previously detected with 301 double haploid lines of the advanced backcross population S42 and (2) to identify new QTLs. Von Korff et al. (2005) detected a total of nine QTLs for powdery mildew and altogether six QTLs for leaf rust resistance. Seven and four QTLs are localized in genomic regions, which are already represented by Hsp introgressions of the evaluated 39 S42ILs. As listed in Table 3, four out of these seven QTLs (57.1%) and three out of these four QTLs (75.0%) were verified with the S42ILs for powdery mildew and leaf rust, respectively. In the S42ILs, the strongest effect reducing the susceptibility to powdery mildew was measured for QPm.S42IL-1H.a on top of chromosome 1H. This effect could be induced through an allele of the Mla resistance locus, which is localized within the same region (Zhou et al. 2001). The QTL QPm.S42IL-4H.b on chromosome 4H, 125-170 cM, which is represented by S42IL-123, showed a significant higher susceptibility than ‘Scarlett’ (42.4%). The region possibly corresponds to the Mlg locus, mapped by Kürth et al. (2001). The Mlg gene is known to be present in ‘Scarlett’, instead of the mlo gene (von Korff et al. 2005). Schönfeld et al. (1996) reported on the Mlf powdery mildew resistance gene, which is located on the short arm of chromosome 7H, close to the QTL QPm.S42IL-7H.a. Here, strong effects reducing the susceptibility to powdery mildew were detected for the S42ILs -135, -137, and -138 (Table 3). The strongest favorable QTL effect for leaf rust (QLr.S42IL-7H.a) was detected for S42IL-138 within the region 7H, 166-181 cM. This effect could possibly be caused by Rph19, a major resistance gene, which was mapped by Park and Karakousis (2002) to a locus, which is only 2 cM distant to QLr.S42IL-7H.a. In future, it is intended to fine-map or sequence the mentioned resistance genes in the corresponding S42ILs to verify the assumption that they indeed are causal for the detected QTL effects.
Bernacchi et al. (1998b) conducted a comprehensive NIL study for tomato to verify QTLs, which were previously detected in two advanced backcross populations (Bernacchi et al. 1998a). They developed a total of 23 NILs, carrying single exotic introgressions either from S. hirsutum or S. pimpinellifolium. The NILs were generated for regions, which contained QTLs with a favorable effect on one or several traits. They covered 15 genomic regions with 25 QTLs for seven agronomic traits. Altogether, 22 QTLs (88%) were confirmed by the NILs. So far, no report is available on the verification of QTL data by a complete or nearly complete barley IL set. However, there are several studies that confirmed QTL effects for powdery mildew resistance genes (Czembor and Czembor 2004) and leaf rust resistance QTLs (van Berloo et al. 2001; Marcel et al. 2007, 2008) by NILs.
Bernacchi et al. (1998b) pointed out that the verification of QTLs with NILs can be characterized with regard to the stability of the QTL effect across environments and the magnitude of the effect. In the present QTL verification study, all four corresponding QTLs for powdery mildew and two out of three corresponding QTLs for leaf rust were significant (P < 0.05) across all environments in the S42 population as well as in the S42ILs. In addition, three and one QTL, respectively, were detected as line × environment interaction effects in the S42ILs. Furthermore, nearly all verified QTLs showed the same direction of the effect in the S42ILs and the S42 population. One exception was the QTL QLr.S42IL-2H.b/QLr.S42-2H.a, which revealed an increased susceptibility to leaf rust in the S42ILs, but a reduction effect in the S42 population. For all QTL effects with the same direction, except QLr.S42IL-4H.a/QLr.S42-4H.a, the magnitude of effect was higher in the S42ILs than in the S42 population. The strongest boost was measured for QPm.S42IL-4H.b which showed an increased effect on powdery mildew susceptibility of 42.4% in S42IL-123, whereas the corresponding QTL QPm.S42-4H.a revealed an effect of 16.1%, relative to ‘Scarlett’ in the S42 population. These discrepancies could be caused by environmental effects since both populations have been tested in different locations and years. In addition, the presence of different strains of powdery mildew and leaf rust in the two experiments could also result in deviating effects. Finally, a reduction in the number of epistatic effects between Hsp alleles, which are present with lower frequency in the S42ILs, can be assumed. Whereas the 301 BC2DH lines of population S42 carry on average four independent exotic introgressions (von Korff et al. 2004), 55 out of the 59 S42ILs contain only single introgressed Hsp segments. To test the epistasis hypothesis we are currently crossing the S42ILs systematically with each other. In future, it is intended to search for epistatic interactions in the resulting plants, each harboring zero, one or two independent Hsp introgressions. With this material, the performance of all four possible allele combinations (Hv/Hv, Hsp/Hsp, Hv/Hsp, and Hsp/Hv) will be compared.
Altogether, three and one new QTL were detected by S42ILs for powdery mildew and leaf rust resistance, respectively. At two QTLs for powdery mildew (QPm.S42IL-1H.b and QPm.S42IL-4H.c), the Hsp introgression caused a reduced susceptibility in comparison to ‘Scarlett’ (Table 3). Three co-localizations of QTL effects for both diseases were exposed by S42ILs on chromosomes 2H, 4H, and 7H (Fig. 3). As illustrated in Table 3, one favorable effect on both diseases was identified by S42IL-138 in the region 7H, 166–181. Due to this, S42IL-138 could be used to improve resistance of current spring barley cultivars to both diseases simultaneously by transfer of Hsp resistance genes. With most of the S42ILs, the QTL effects could be precisely localized to one chromosomal region, since they only carry one target introgression. In contrast, the S42ILs -102, -114, -123, and -129 each contain one additional non-target Hsp segment (Table 1). Due to this, a precise QTL mapping with these lines is not possible so far. It can be assumed that (1) only one introgression, either the target or the non-target one, causes a significant QTL effect or that (2) there are two independent QTL effects, localized in both the target and the non-target introgressions.
There are several instances where a S42IL revealed a significant QTL effect, whereas overlapping sister lines containing Hsp alleles in the same chromosomal region do not show this effect. These discrepancies can be attributed to the following three reasons: (1) the true size of an exotic introgression might slightly differ from our estimation which is based on genotype data from a limited number of 98 SSR markers (see Fig. 3). Thus, it might be possible that Hsp alleles, present in an extended introgression, cause a QTL effect, which is missing in the apparent sister S42IL. For example, S42IL-108, revealing no QTL effect, might actually possess the estimated introgression 2H, 17-92 cM. However, the sister line S42IL-107 (2H, 17–42 cM), harboring a QTL for powdery mildew, might eventually contain an introgression which begins further north of the first chromosomal SSR marker (2H, 17 cM), resulting in a unique QTL effect which is not detectable in the sister line S42IL-108. (2) As an alternative explanation, the IL with the larger introgression could include additional Hsp alleles which interact with the QTL present in the small introgression. This interaction could result in a loss of significant QTL effects. Under this light, a second Hsp allele could be present in S42IL-108, located south of the small introgression in S42IL-107, which counter-acts or turns down the original QTL effect. (3) Some S42ILs might also contain a so far hidden non-target introgression, carrying a Hsp allele which exhibit the significant QTL effect. This explanation could eventually be the case for S42IL-110 on chromosome 2H, 80-86 cM, where two QTL effects are present. In contrast, no QTL was detected in the sister lines S42IL-108 and -109 although the latter two ILs completely cover the introgression of the former IL. To test the three hypotheses presented, it is required to characterize the introgressions more precisely with additional DNA markers in order (1) to define the extensions of the introgressions with a higher resolution, (2) to discover and then eliminate potential extra non-target introgressions, and (3) to fine-map and possibly dissect the detected QTL effects in sub-ILs carrying smaller Hsp segments.
In future, there are several demands concerning the further development and evaluation of the S42IL set. The first aim will be the completion of the IL set by selecting lines for the chromosomal Hsp regions, which are so far not represented. As mentioned above, it is also intended to increase the marker density by genotyping new DNA markers. In addition, Sub-ILs will be selected and used for high-resolution mapping as a first step toward map-based cloning of interesting QTL. The S42ILs will also be extensively characterized on phenotype level. Here, the focus will be laid on agronomic traits, malting quality, biotic, and abiotic stress. All genotype and phenotype data of the S42ILs will be archived for general use in the public IL data base ‘Phenom Networks’ (http://phn.huji.ac.il/RTQ/). Our first application demonstrates that the S42ILs are a powerful genetic tool to unravel the genetic architecture of agriculturally relevant traits, as well as for providing cereal breeders with newly selected exotic germplasm.
References
Aida Y, Tsunematsu H, Doi K, Yoshimura A (1997) Development of a series of introgression lines of Japonica in the background of Indica rice. Rice Genet Newsl 14:41–43
Badr A, Müller K, Schäfer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510
Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick PW, Fernie AW, Sweetlove LJ (2005) Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant Cell Physiol 46:425–437
Ben Chaim A, Borovsky Y, Rao GU, Tanyolac B, Paran I (2003) fs3.1: a major fruit shape QTL conserved in Capsicum. Genome 46:1–9
Bernacchi D, Beck-Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S (1998a) Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397
Bernacchi D, Beck-Bunn T, Emmatty D, Eshed Y, Inai S, Lopez J, Petiard V, Sayama H, Uhlig J, Zamir D, Tanksley S (1998b) Advanced back-cross QTL analysis of tomato. II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon hirsutum and L. pimpinellifolium. Theor Appl Genet 97:1191–1196
Brown AHD, Munday J, Oram RN (1988) Use of isozyme-marked segments from wild barley (Hordeum spontaneum) in barley breeding. Plant Breed 100:280–288
Canady MA, Meglic V, Chetelat RT (2005) A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48:685–697
Concibido VC, La Vallee B, Mclaird P, Pineda N, Meyer J, Hummel L, Yang J, Wu K, Delannay X (2003) Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor Appl Genet 106:575–582
Czembor PC, Czembor JH (2004) DNA polymorphism among barley NILs of cv. Pallas, carrying genes for resistance to powdery mildew (Blumeria graminis f. sp. hordei). J Appl Genet 45:183–187
Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121
Eduardo I, Arus P, Monforte AJ (2005) Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor Appl Genet 112:139–148
Eduardo I, Arus P, Monforte AJ, Obando J, Fernandez-Trujillo JP, Martinez JA, Alarcon AL, Alvarez JM, van der Knaap E (2007) Estimating the genetic architecture of fruit quality traits in melon using a genomic library of near isogenic lines. J Am Soc Hortic Sci 132:80–89
Eshed Y, Abu-Abied M, Saranga Y, Zamir D (1992) Lycopersicon esculentum lines containing small overlapping introgressions from Lycopersicon pennellii. Theor Appl Genet 83:1027–1034
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in Lycopersicon esculentum—a tool for fine mapping of genes. Euphytica 79:175–179
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Fernandez-Trujillo JP, Obando J, Martinez JA, Alarcon AL, Eduardo I, Arus P, Monforte AJ (2007) Mapping fruit susceptibility to postharvest physiological disorders and decay using a collection of near-isogenic lines of melon. J Am Soc Hortic Sci 132:739–748
Finkers R, van Heusden AW, Meijer-Dekens F, van Kan JAL, Maris P, Lindhout P (2007) The construction of a Solanum habrochaites LYC4 introgression line population and the identification of QTLs for resistance to Botrytis cinerea. Theor Appl Genet 114:1071–1080
Frary A, Nesbitt TC, Frary A, Grandillo S, van der Knaap E, Cong B, Liu JP, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305:1786–1789
Fulton TM, Nelson JC, Tanksley SD (1997) Introgression and DNA marker analysis of Lycopersicon peruvianum, a wild relative of the cultivated tomato, into Lycopersicon esculentum, followed through three successive backcross generations. Theor Appl Genet 95:895–902
Garcia GM, Stalker HT, Kochert G (1995) Introgression analysis of an interspecific hybrid population in peanuts (Arachis hypogaea L.) using RFLP and RAPD marker. Genome 38:166–176
Hanson PM, Sitathani K, Sadashiva AT, Yang RY, Graham E, Ledesma D (2007) Performance of Solanum habrochaites LA 1777 introgression line hybrids for marketable tomato fruit yield in Asia. Euphytica 158:167–178
Holtan HE, Hake S (2003) Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon penellii segmental introgression lines. Genetics 165:1541–1550
Hori K, Sato K, Nankaku N, Takeda K (2005) QTL analysis in recombinant chromosome substitution lines and doubled haploid lines derived from a cross between Hordeum vulgare ssp. vulgare and Hordeum vulgare ssp. spontaneum. Mol Breed 16:295–311
Howell PM, Marshall DF, Lydiate DJ (1996) Towards developing intervarietal substitution lines in Brassica napus using marker-assisted selection. Genome 39:348–358
Jena KK, Khush GS, Kochert G (1992) RFLP analysis of rice (Oryza sativa L.) introgression lines. Theor Appl Genet 84:608–616
Jeuken MJW, Lindhout P (2004) The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor Appl Genet 109:394–401
Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Vries HBD, Effgen S, Vreugdenhil D, Koornneef M (2007) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175:891–905
Kürth J, Kolsch R, Simons V, Schulze-Lefert P (2001) A high-resolution genetic map and a diagnostic RFLP marker for the Mlg resistance locus to powdery mildew in barley. Theor Appl Genet 102:53–60
Leonova IN, Laikova LI, Popova OM, Unger O, Börner A, Röder MS (2007) Detection of quantitative trait loci for leaf rust resistance in wheat-T.timopheevii/T.tauschii introgression lines. Euphytica 155:79–86
Li ZK, Fu BY, Gao YM, Xu JL, Ali J, Lafitte HR, Jiang YZ, Rey JD, Vijayakumar CHM, Maghirang R, Zheng TQ, Zhu LH (2005) Genome-wide introgression lines and their use in genetic and molecular dissection of complex phenotypes on rice (Oryza sativa L.). Plant Mol Biol 59:33–52
Lippman ZB, Semel Y, Zamir D (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Gen & Dev 17:545–552
Liu SB, Zhou RG, Dong YC, Li P, Jia JZ (2006) Development, utilization of introgression lines using a synthetic wheat as donor. Theor Appl Genet 112:1360–1373
Liu SW, Zhao SY, Chen F, Xia GM (2007) Generation of novel high quality HMW-GS genes in two introgression lines of Triticum aestivum/Agropyron elongatum. BMC Evol Biol 7:76
Marcel TC, Aghnoum R, Durand J, Varshney RK, Niks RE (2007) Dissection of the barley 2L1.0 region carrying the ‘Laevigatum’ quantitative resistance gene to leaf rust using near-isogenic lines (NIL) and subNIL. Mol Plant Microbe Interact 20:1604–1615
Marcel TC, Gorguet B, Ta MT, Kohutova Z, Vels A, Niks RE (2008) Isolate specificity of quantitative trait loci for partial resistance of barley to Puccinia hordei confirmed in mapping populations and near-isogenic lines. New Phytol 177:743–755
Matus I, Corey A, Filichkin T, Hayes PM, Vales MI, Kling J, Riera-Lizarazu O, Sato K, Powell W, Waugh R (2003) Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp spontaneum as a source of donor alleles in a Hordeum vulgare subsp vulgare background. Genome 46:1010–1023
Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Ying CS, Luo LJ (2006) QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor Appl Genet 112:648–656
Monforte AJ, Tanksley SD (2000) Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L.esculentum genetic background: A tool for gene mapping and gene discovery. Genome 43:803–813
Obando J, Fernandez-Trujillo JP, Martinez JA, Alarcon AL, Eduardo I, Arus P, Monforte AJ (2008) Identification of melon fruit quality quantitative trait loci using near-isogenic lines. J Am Soc Hortic Sci 133:139–151
Overy A, Walker HJ, Malone S, Howard TP, Baxter CJ, Sweetlove LJ, Hill SA, Quick WP (2005) Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J Exp Bot 56:287–296
Park RF, Karakousis A (2002) Characterization and mapping of gene Rph19 conferring resistance to Puccinia hordei in the cultivar ‘Reka 1’ and several Australian barleys. Plant Breed 121:232–236
Percy RG, Cantrell RG, Zhang J (2006) Genetic variation for agronomic and fiber properties in an introgressed recombinant inbred population of cotton. Crop Sci 46:1311–1317
Pestsova EG, Börner A, Röder MS (2006) Development and QTL assessment of Triticum aestivum–Aegilops tauschii introgression lines. Theor Appl Genet 112:634–647
Prakash NS, Combes MC, Somanna N, Lashermes P (2002) AFLP analysis of introgression in coffee cultivars (Coffea arabica L.) derived from a natural interspecific hybrid. Euphytica 124:265–271
Ramsay LD, Jennings DE, Bohuon EJR, Arthur AE, Lydiate DJ, Kearsey MJ, Marshall DF (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39:558–567
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97:11102–11107
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, Graner A, Close TJ, Waugh R (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. PNAS 103:18656–18661
Rousseaux MC, Jones CM, Adams D, Chetelat R, Bennett A, Powell A (2005) QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor Appl Genet 111:1396–1408
Röder MS, Huang XQ, Börner A (2008) Fine-mapping of the region on wheat chromosome 7D controlling grain weight. Funct Integr Genomics 8:79–86
SAS Institute (2006) The SAS Enterprise Guide for Windows, release 4.1. SAS Institute, Cary
Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotech 24:447–454
Schönfeld M, Ragni A, Fischbeck G, Jahoor A (1996) RFLP mapping of three new loci for resistance genes to powdery mildew (Erysiphe graminis f sp hordei) in barley. Theor Appl Genet 93:48–56
Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103:12981–12986
Siangliw L, Jongdee B, Pantuwan G, Toojinda T (2007) Developing KDML105 backcross introgression lines using marker-assisted selection for QTLs associated with drought tolerance in rice. Sci Asia 33:207–214
Simón MR, Ayala FM, Cordo CA, Röder MS, Börner A (2007) The use of wheat/goatgrass introgression lines for the detection of gene(s) determining resistance to septoria tritici blotch (Mycosphaerella graminicola). Euphytica 154:249–254
Song W, Xie H, Liu Q, Xie CJ, Ni ZF, Yang TM, Sun QX, Liu ZY (2007) Molecular identification of Pm12-carrying introgression lines in wheat using genomic and EST-SSR markers. Euphytica 158:95–102
Szalma SJ, Hostert BM, LeDeaux JR, Stuber CW, Holland JB (2007) QTL mapping with near-isogenic lines in maize. Theor Appl Genet 114:1211–1228
Tan LB, Liu FX, Xue W, Wang GJ, Ye S, Zhu ZF, Fu YC, Wang XK, Sun CQ (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J Integr Plant Biol 49:871–884
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, Sun CQ (2006a) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet 112:570–580
Tian F, Zhu ZF, Zhang BS, Tan LB, Fu YC, Wang XK, Sun CQ (2006b) Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.). Theor Appl Genet 113:619–629
van Berloo R, Aalbers H, Werkman A, Niks RE (2001) Resistance QTL confirmed through development of QTL–NILs for barley leaf rust resistance. Mol Breed 8:187–195
van der Knaap E, Sanyal A, Jackson SA, Tanksley SD (2004) High-resolution fine mapping and fluorescence in situ hybridization analysis of sun, a locus controlling tomato fruit shape, reveals a region of the tomato genome prone to DNA rearrangements. Genetics 168:2127–2140
von Korff M, Wang H, Léon J, Pillen K (2004) Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor Appl Genet 109:1736–1745
von Korff M, Wang H, Léon J, Pillen K (2005) AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor Appl Genet 111:583–590
von Korff M, Wang H, Léon J, Pillen K (2006) AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 112:1221–1231
von Korff M, Wang H, Léon J, Pillen K (2008) AB-QTL analysis in spring barley: III. Identification of exotic alleles for the improvement of malting quality in spring barley (H. vulgare ssp. spontaneum). Mol Breed 21:81–93
Wenzl P, Li HB, Carling J, Zhou MX, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesna J, Cakir M, Poulsen D, Wang JP, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics 7:206
Xu JL, Lafitte HR, Gao YM, Fu BY, Torres R, Li ZK (2005) QTLs for drought escape and tolerance identified in a set of random introgression lines of rice. Theor Appl Genet 111:1642–1650
Yates HE, Frary A, Doganlar S, Frampton A, Eannetta NT, Uhlig J, Tanksley SD (2004) Comparative fine mapping of fruit quality QTLs on chromosome 4 introgressions derived from two wild tomato species. Euphytica 135:283–296
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nature Rev Genet 2:983–989
Zhang X, Zhou SX, Fu YC, Su Z, Wang XK, Sun CQ (2006) Identification of a drought tolerant introgression line derived from dongxiang common wild rice (O. rufipogon griff.). Plant Mol Biol 62:247–259
Zheng TQ, Xu JL, Li ZK, Zhai HQ, Wan JM (2007) Genomic regions associated with milling quality and grain shape identified in a set of random introgression lines of rice (Oryza sativa L.). Plant Breed 126:158–163
Zhou FS, Kurth JC, Wei FS, Elliott C, Vale G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P (2001) Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13:337–350
Zhou SX, Tian F, Zhu ZF, Fu YC, Wang XK, Sun CQ (2006) Identification of quantitative trait loci controlling drought tolerance at seedling stage in Chinese Dongxiang common wild rice (Oryza rufipogon Griff.). Acta Genetica Sinica 33:551–558
Acknowledgments
We are indebted to Prof. J. Léon for providing the experimental field station Dikopshof, University of Bonn, for field experiments. We thank Drs. E. Laubach (Nordsaat Saatzucht), J. Breun and H. Kempe (Saatzucht Josef Breun) and W. Bungert and H. Rehkopf (Dikopshof) and their teams for conducting the field experiments. The excellent technical assistance of M. Noschinski is appreciated. This work was funded by the German Plant Genome Research Initiative (GABI) of the Federal Ministry of Education and Research (BMBF, project 0313125B).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. J. Muehlbauer.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Schmalenbach, I., Körber, N. & Pillen, K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet 117, 1093–1106 (2008). https://doi.org/10.1007/s00122-008-0847-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0847-7