Abstract
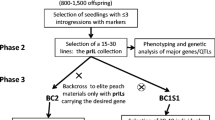
Backcross inbred lines (BILs) were developed in which chromosome segments of Lactuca saligna (wild lettuce) were introgressed into L. sativa (lettuce). These lines were developed by four to five backcrosses and one generation of selfing. The first three generations of backcrossing were random. Marker-assisted selection began in the BC4 generation and continued until the final set of BILs was reached. A set of 28 lines was selected that together contained 96% of the L. saligna genome. Of these lines, 20 had a single homozygous introgression (BILs), four had two homozygous introgressions (doubleBILs) and four lines had a heterozygous single introgression (preBILs). Segregation ratios in backcross generations were compared to distorted segregation ratios in an F2 population, and the results indicated that most of the distorted segregations can be explained by genetic effects on pollen- or egg-cell fitness. By means of BIL association mapping we were able to map 12 morphological traits and hundreds of additional amplified fragment length polymorphic (AFLP) markers. The total AFLP map now comprises 757 markers. This set of BILs is very useful for future genetic studies.




Similar content being viewed by others
References
Bonnier FJM, Reinink K, Groenwold R (1992) New sources of major gene resistance in Lactuca to Bremia lactucae. Euphytica 61:203–211
Chetelat RT, Meglic V (2000) Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor Appl Genet 100:232–241
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79:175–179
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Grandillo S, Ku HM, Tanksley SD (1999) Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor Appl Genet 99:978–987
Gustafsson I (1989) Potential sources of resistance to lettuce downy mildew (Bremia lactucae) in different Lactuca species. Euphytica 40:227–232
Jeuken M, Lindhout P (2002) Lactuca saligna, a non-host for lettuce downy mildew (Bremia lactucae), harbors a new race-specific Dm gene and three QTLs for resistance. Theor Appl Genet 105:384–391
Jeuken M, van Wijk R, Peleman P, Lindhout P (2001) An integrated interspecific AFLP map of lettuce (Lactuca) based on two L. sativa × L. saligna F2 populations. Theor Appl Genet 103:638–647
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lebeda A, Boukema IW (1991) Further investigation of the specificity of Interactions between wild Lactuca spp. and Bremia lactucae isolates from Lactuca serriola. J Phytopathol 133:57–64
Lin SY, Yamamoto T, Sato M, Shomura A, Shimano T, Kuboki Y, Harushima Y, Yano M, Lin SY (1996) Backcross inbred lines as a permanent mapping population, Rice Genome 5, issue 1
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines, Theor Appl Genet 96:997–1003
Lindqvist K (1956) Reflexed and erect involucre in Lactuca. Hereditas 42:436–442
Lindqvist K (1960) Inheritance studies in lettuce. Hereditas 46:387–470
Monforte AJ, Tanksley SD (2000) Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43:803–813
Monforte AJ, Tanksley SD (2001) Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor Appl Genet 100:471–479
Norwood JM, Crute IR, Lebeda A (1981) The location and characteristics of novel sources of resistance to Bremia lactucae Regel (downy mildew) in wild Lactuca species. Euphytica 30:659–668
Robinson RW, McCreight JD, Ryder EJ (1983) The genes of lettuce and closely related species. Plant Breed Rev 1:267–293
Van Berloo R (1999) The development of software for the graphical representation and filtering of molecular marker data: graphical genotypes (GGT). J Hered 90:328–329
Van Berloo R, Lindhout P (2001) Mapping disease resistance genes in tomato. In: Zhu D, Hawtin G, Wang Y (eds) Int Symp Biotechnol Applic Hortic Crops, vol 12. China Agricultural Scientech Press, Beijing, pp 343–356
Van der Beek JG, Verkerk R, Zabel P, Lindhout P (1992) Mapping strategy for resistance genes in tomato based on RFLPs between cultivars: Cf-9 (resistance to Cladosporium fulvum) on chromosome 1. Theor Appl Genet 84:106–112
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Young ND (1996) QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol 34:479–501
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989
Acknowledgements
We thank P. Stam and R.E. Niks for critically reading the manuscript and for their advisory support in this study. We are thankful for the financial and advisory support of the Dutch lettuce breeding companies.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Salamini
Rights and permissions
About this article
Cite this article
Jeuken, M.J.W., Lindhout, P. The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor Appl Genet 109, 394–401 (2004). https://doi.org/10.1007/s00122-004-1643-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1643-7