Abstract
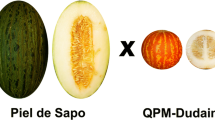
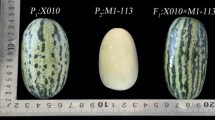
A doubled haploid line (DHL) population of melon derived from a cross between the Korean cultivar “Songwhan Charmi” accession PI161375 (SC), included in the horticultural group conomon, and the Spanish cultivar “Piel de Sapo” (PS), included in the horticultural group inodorus, was used to develop a collection of near isogenic lines (NILs). These parental lines represent very different melon cultivar groups, with important differences at fruit, plant, disease response and molecular level. This cross is one of the most polymorphic ones within melon germplasm. Selected DHLs were backcrossed to PS and further backcrossing and selfing was performed, monitoring introgressions from SC using molecular markers covering the melon genetic map. A final collection of 57 NILs was obtained, containing a unique independent introgression from SC in the PS genetic background. The introgressions within the collection cover at least 85% of the SC genome with an average introgression size of 41 cM, corresponding to 3.4% of the SC genome. The average resolution for mapping genes or quantitative trait loci is 18.90 cM. This set of NILs is a potentially powerful tool for the study of quantitative trait locus involved in melon fruit quality and other important complex traits, and the introduction of new genetic variability in modern cultivars from exotic sources. The NILs can also be used as pre-competitive breeding lines in melon breeding projects.




Similar content being viewed by others
References
Akashi Y, Fukunda N, Wako T, Masuda M, Kato K (2002) Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica 125:385–396
Aranzana MJ, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriano G, Ryder CD, Testolin R, Abbott A, King GJ, Iezzoni F, Arús P (2003) A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor Appl Genet 106:819–825
Bernacchi D, Tanksley SD (1997) An interspecific backcross of Lycopersicon esculentum x L. hirsutum: Linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147:861–877
Bernacchi D, Beck-Bunn T, Emmatty D, Eshed J, Inai S, Lopez J, Petiard V, Sayama H, Uhlig J, Zamir D, Tanksley SD (1998) Advanced backcross QTL analysis of tomato II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon hirsutum and L. pimpinellifolium. Theor Appl Genet 97:170–180
Chen JF, Adelberg J (2000) Interspecific hybridization in Cucumis-Progress, problems, and perspectives. Hortscience 35:11–14
Chetelat RT, Meglic V (2000) Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor Appl Genet 100:232–241
Eduardo I, Arús P, Monforte AJ (2004) Genetics of fruit quality in melon. Verification of QTLs involved in fruit shape with near-isogenic lines (NILs). In: Lebeda A, Paris S (eds) Progress in cucurbit genetics and breeding research. Palacký University, Olomouc, pp 499–502
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79:175–179
Eshed Y, Zamir D (1995) An introgression-line population of Lycopersicon pennelli in the cultivated tomato enables the identification and fine mapping of yield-associated QTLs. Genetics 141:1147–1162
Falconer DS (1989) Introduction to quantitative genetics, 3rd edn. Longman scientific & technical, England
Frary A, Nesbitt TC, Grandillo S, Van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97:4718–4723
Fukino N, Kusuya M, Kunihisa M, Matsumoto S (2004) Characterization of simple sequence repeats (SSRs) and developemnt of SSR markers in melon (Cucumis melo). In: Lebeda A, Paris S (eds) Progress in cucurbit genetics and breeding research. Palacký University, Olomouc, pp 503–506
Fulton TM, Beck-Bunn T, Emmatty D, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley SD (1997) QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor Appl Genet 95:881–894
Garcia-Mas J, Oliver M, Gómez-Paniagua H, de Vicente MC (2000) Comparing AFLP, RAPD and RFLP markers for measuring genetic diversity in melon. Theor Appl Genet 101:860–864
Gonzalo MJ (2003) Generación, caracterización molecular y evaluación morfológica de una población de líneas dihaploides en melón (Cucumis melo L.). PhD Dissertation, Universitat de Lleida, Spain
Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arús P Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811
Hyne V, Kearsey MJ, Pike DJ, Snape JW (1995) QTL analysis: unreliability and bias in estimation procedures. Mol Breed 1:273–282
Jeuken MJW, Lindhout P (2004) The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor Appl Genet 109:394–401
Kearsey MJ, Farquhar AGL (1998) QTL analysis in plants: where are we now? Heredity 80:137–142
Kirkbride JH (1993) Biosystematic monograph of the genus Cucumis (Cucurbitaceae). Parkway Publishers, North Caroline
Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3(3):493–494
Koumproglou R, Wilkes TW, Townson P, Wang XY, Beynon J, Pooni HS, Newbury HJ, Kearsey MJ (2002) STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J 31(3):355–364
Lebreton CH, Visscher PM, Haley CS, Semikhodskii A, Quarrie SA (1998) A nonparametric bootstrap method for testing close linkage vs. pleiotrophy of coincident quantitative trait loci. Genetics. 150:931–943
Liu J, van Eck J, Cong B, Tanksley (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99:13302–13306
Liu L, Kakihara F, Kato M (2004) Characterization of six varieties of Cucumis melo L. based on morphological and physiological characters, including shelf-life of fruit. Euphytica 135:305–313
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Mackay TF (2001) The genetic architecture of quantitative traits. Annu Rev Genet 35:303–339
Melchinger AE, Utz HF, Schön CC (1998) Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149:383–403
Mliki A, Staub JE, Sun ZY, Ghorbel A (2001) Genetic diversity in melon (Cucumis melo L.): An evaluation of African germplasm. Genet Resour Crop Evol 48(6):587–597
Monforte AJ, Tanksley SD (2000a) Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43:803–813
Monforte AJ, Tanksley SD (2000b) Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor Appl Genet 100:471–479
Monforte AJ, Friedman E, Zamir D, Tanksley SD (2001) Comparison of a set of allelic QTL-NILs for chromosome 4 of tomato: deductions about natural variation and implications for germplasm utilization. Theor Appl Genet 102:572–590
Monforte AJ, Garcia-Mas J, Arús P (2003) Genetic variability in melon based on microsatellite variation. Plant Breed 122:1–6
Monforte AJ, Oliver M, Gonzalo MJ, Álvarez JM, Dolçet-Sanjuan R, Arús P (2004) Identification of quantitative trait loci involved in fruit quality traits in melon. Theor Appl Genet 108:750–758
Monforte AJ, Eduardo I, Abad S, Arús P (2005) Inheritance mode of fruit traits in melon. Heterosis for fruit shape and its correlation with genetic distance. Euphytica 144:31–38
Morales M, Roig E, Monforte AJ, Arús P, Garcia-Mas J (2004) Single-nucleotide polymorphisms detected in expressed sequence tags of melon (Cucumis melo L.). Genome 47:352–360
Morgante M, Salamini F (2003) From plant genomics to breeding practice. Curr Opin Biotech 14:214–219
Oliver M, Garcia-Mas J, Cardús M, Pueyo N, López-Sesé AI, Arroyo M, Gómez-Paniagua H, Arús P, de Vicente MC (2001) Construction of a reference linkage map for melon. Genome 44:836–845
Overy SA, Walker HJ, Malone S, Howard TP, Baxter CJ, Sweetlove LJ, Hill SA, Quick WP (2004) Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J Exp Bot 56:287–296
Paran I, Zamir D (2003) Quantitative traits in plants: beyond the QTL. Trends Genet 19(6):303–306
Paterson AH, DeVerna JW, Lanini B, Tanksley SD (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124:735–742
Périn C, Hagen LS, Giovinazzo N, Besombes D, Dogimont C, Pitrat M (2002a) Genetic control of fruit shape acts prior to anthesis in melon (Cucumis melo L.). Mol Gen Genomics 266:933–941
Périn C, Hagen L S, De Conto V, Katzir N, Danin-Poleg Y, Portnoy V, Baudracco-Arnas S, Chadoeuf J, Dogimont C, Pitrat M (2002b) A reference map of Cucumis melo based on two recombinant inbred lines populations. Theor Appl Genet 104:1017–1034
Pestsova EG, Borner A, Roder MS (2001) Development of a set of Triticum aestivum–Aegilops tauschii introgression lines. Hereditas 135(2–3):139–143
Pillen K, Ganal MW, Tanksley SD (1996) Construction of a high-resolution genetic map and YAC-contigs in the tomato Tm-2a region. Theor Appl Genet 93:228–233
Puigdomènech P, Martínez-Izquierdo JA, Arús P, Garcia-Mas J, Monforte AJ, Nuez F, Picó B, Blanca J, Aranda M, Arnau V, Robles A (2005) The Spanish melon genomics initiative
Ramsay LD, Jennings DE, Bohuon EJR, Arthur AE, Lydiate DJ, Kearsey MJ, Marshal DF (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39:558–567
Ritschel PS, de Lima Lins TC , Tristan RL , Buso GSC , Buso JA, Ferreira ME (2004) Development of microsatellite markers from an enriched genomic library for genetic analysis of melon (Cucumis melo L.). BMC Plant Biol 4:9. DOI:10.1186/1471–2229–4–9
Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH (2004) Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304:445–448
Stepansky A, Kovalski I, Perl-Treves R (1999) Intraspecific classification of melons (Cucumis melo L.) in view of their phenotypic and molecular variation. Plant Syst Evol 217:313–332
Tanksley SD (1993). Mapping polygenes. Annu Rev Genet 27:205–233
Tanksley SD, Grandillo S, Fulton TM, Zamir D, Eshed T, Petiard V, Lopez J, Beck-Bunn T (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor Appl Genet 92:213–224
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Van Berloo R (1999) The development of software for the graphical representation and filtering of molecular marker data: graphical genotypes (GGT). J Hered 90:328–329
von Korff M, Wang H, León J, Pillen K (2004) Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor Appl Genet 109:1736–1745
Wan XY, Wan JM, Su CC, Wang CM, Shen WB, Li JM, Wang HL, Jiang L, Liu SJ, Chen LM, Yasui H, Yoshimura A (2004) QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theor Appl Genet 110:71–79
Wehrhahn C, Allard R W (1965) The detection and measurement of the effects of individual genes involved in the inheritace of a quantitative character in wheat. Genetics 51:109–119
Whitaker TW, Davis GN (1962) Cucurbits, botany, cultivation and utilization. Interscience Pub, New York
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2: 983–989
Acknowledgements
The authors thank N. Galofré, I. Marchal, A. Montejo A. Ortigosa, J. Adillón and P. Ramon for technical assistance. This work was funded in part by grants AGL2000–0360 and AGL2003–09175-C02–01, from the Spanish Ministry of Education and Science. AJM was partly supported by a contract from Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA). IE was supported by a fellowship from the Spanish Ministry of Education. The experiments presented here comply with current Spanish law.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran
Rights and permissions
About this article
Cite this article
Eduardo, I., Arús, P. & Monforte, A.J. Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor Appl Genet 112, 139–148 (2005). https://doi.org/10.1007/s00122-005-0116-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0116-y