Abstract
Background
The intestinal microbiota play remarkable roles in maintaining the health of their hosts. Recent studies focused on gut bacterial diversity in birds and poultry, with little information about the ecological functions of their gut fungal community.
Methods
The high-throughput sequencing was applied to compare intestinal fungal community structure between Hooded Crane (Grus monacha) and Domestic Goose (Anser anser domesticus), and infer the potential pathogens of each species at Shengjin Lake of China.
Results
Intestinal fungal alpha diversity was higher in Hooded Crane than Greylag Goose (Anser anser). Gut fungal community composition showed dramatic shifts between the two species. Hooded Cranes mainly eat Vallisneria natans and Potamogeton malaianus, while artificial hurl food (i.e., paddy) was the main food resource for Domestic Geese, suggesting that the variations in fungal community might be induced by different diets between the two hosts. Two enriched genera (i.e., Acremonium and Rhodotorula) which could increase host’s digestion were detected in guts of Hooded Cranes. In addition, there were 42 pathogenic amplicon sequence variants (ASVs), 17% of which shared in Hooded Crane and Greylag Goose. The Hooded Crane had higher gut fungal pathogenic diversity and abundance relative to Greylag Goose.
Conclusions
The study demonstrated that divergence in intestinal fungal community structure might be induced by different diets between wintering Hooded Crane and Domestic Goose. Hooded Crane might rely more on their gut fungal taxa to acquire nutrients from indigestible food resources. Our study also implied that more research should focus on intestinal pathogens in wild birds and domestic poultry, as they might increase risk of disease in other animals, even human beings. The degree of cross infection in pathogens among wild birds and sympatric poultry should be clearly verified in future study.
Similar content being viewed by others
Background
Empirical studies have demonstrated that gut microbiota play crucial roles for their hosts (Xiang et al. 2019), such as helping digestion (Stanley et al. 2012), improving immunity (Chung et al. 2012) and regulating metabolism (Liu et al. 2020). The intestinal microbiota may even affect hosts’ behaviors (e.g. mate choice; Sharon et al. 2010). There might be many factors influenced animal intestinal microbial community patterns, including diet (Bolnick et al. 2014; Fu et al. 2020), life style (Nicholson et al. 2012), genotype (Eckburg et al. 2005; Xiang et al. 2019), gender (Xu et al. 2016; Mason et al. 2019), and seasonal fluctuations (Dong et al. 2019; Zhang et al. 2020).
Migratory birds exhibit unique life history, representing an interesting study object for their gut microbiota. However, there is less information about gut microbiota of wild migratory birds. Hooded Crane (Grus monacha) is a long-distance migratory wild bird, defined as Threatened Species in the International Union for Conservation of Nature and Natural Resources (IUCN) Red List and the first-class national protected wild animal in China. Hooded Crane mainly breeds in south Siberia. From October to next April, they fly to China, Japan and South Korea for wintering (Zheng et al. 2015). Recent studies focused on gut bacterial community structure of Hooded Crane, showing that seasonal dynamics significantly affected gut bacterial diversity (Zhang et al. 2020). Gut bacterial community structure showed dramatic shifts between Hooded Crane and sympatric goose (Xiang et al. 2019; Fu et al. 2020). However, little is known about their gut fungal community. Gut fungi increased nutrition levels for their hosts (Tanahashi et al. 2010, 2017). The roles of gut fungi include provisioning of essential amino acids (Ayayee et al. 2016) and contributing to lignocellulose degradation (Geib et al. 2008; Scully et al. 2012; Herr et al. 2016). Thus, understanding intestinal fungal community might be important for clarifying their ecological function in helping digestion for their hosts.
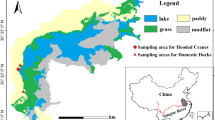
The Shengjin Lake, an internationally important wetland, is a river-connected shallow lake in the middle of the Yangtze River floodplain. The Shengjin Lake is an important wetland for migratory birds, as it can offer suitable feeding habitats in wintering period. However, the lake is facing serious degradation by anthropic activity in recent years, decreasing food availability for wintering birds (Yang et al. 2015). The Hooded Crane changed their dietary structure and foraged together with poultry in paddy lands (Fu et al. 2020). There are large number of Domestic Goose (Anser anser domesticus; poultry) in this region. The overlap of foraging niche between Hooded Crane and Greylag Goose (Anser anser) might increase the transmission of their gut microbiota and pathogens.
Hooded Cranes fly long distance and live various habitats, thus they might contact a large range of pathogens. Intestinal pathogens could lead to series diseases in wild birds (Singh et al. 2013). Previous studies have demonstrated that different hosts could mutually transmit intestinal pathogens in an overlapping environment through physical contact, air, water, soil, food, or other media (Alm et al. 2018; Fu et al. 2020). Thus, those pathogens in Hooded Cranes could spread to conspecifics and/or sympatric poultry, and vice versa (Altizer et al. 2011). However, the assumption of the cross infection of pathogens between migratory birds and poultry has not been verified. In this study, high-throughput sequencing method (Illumina MiSeq) was used to compare the gut fungal community structure between wintering Hooded Crane and sympatric Greylag Goose, and infer the potential fungal pathogens of each species at the Shengjin Lake.
Methods
Sample collection
The Shengjin Lake is a river-connected lake, serving as indispensable wintering and stopover habitat for migratory birds on the East Asia-Australasian flyway (Fox et al. 2011). Hooded Cranes and Greylay Geese often foraged together in paddy fields (Fu et al. 2020). Hooded Crane mainly eats Vallisneria natans and Potamogeton malaianus (Zheng et al. 2015). Artificial hurl food (i.e., paddy) was the main food resource for Domestic Goose. Fecal samples of Hooded Crane and Greylag Goose were collected on December 10, 2018 at the Shegan region, Shengjin Lake. Before sampling, we searched the flocks of Hooded Crane and adjacent Greylag Goose in paddy land. The fresh fecal samples of Hooded Cranes were collected after foraging. The distance among fecal samples was > 5 m to avoid individual repetition. The fecal samples of Domestic Geese (i.e., these geese had overlap foraging niche with Hooded Cranes in paddy field) were collected after artificial hurl food in the yard of farmer’s house. The fecal samples were immediately transported to the lab (i.e., within 12 h) and stored at − 20 °C.
The DNA extraction
Fecal DNA was extracted by the Qiagen DNA Stool Mini Kit according to the DNA isolation protocol. The extracted DNA was dissolved in 60 μL of elution buffer, quantified by NanoDrop ND-1000 (Thermo Scientific, USA), and stored at − 20 °C.
Bird species determination
The COL gene was amplified with primer BIRDF1/BIRDR1 (Hebert et al. 2004). The detailed information about PCR reaction was showed in our previous study (Xiang et al. 2019). The amplicon was sequenced and blasted (> 99% identity) in National Center for Biotechnology Information (NCBI) to identify bird species. We only kept these samples belonged to Hooded Crane for the downstream analysis. The fecal samples of Domestic Goose were collected under certain circumstance (i.e., without disturbance of other species), so we need not perform determination for Domestic Goose.
PCR and amplicon library preparation
A total of 36 fecal samples (each species with 18 samples) were used in this study. PCR was performed using primer ITS1/ITS2 in 50 μL mixtures with parameters as follows: 35 cycles of denaturation at 95 °C for 45 s, annealing at 56 °C for 45 s, and extension at 72 °C for 45 s; with a final extension at 72 °C for 10 min. The PCR products were purified for sequencing.
Bioinformatics and statistical analysis
Fungal raw data were processed by QIIME (v2-2020.2; Bolyen et al. 2019). The deblur algorithm was used to filter the low quality sequences (Amir et al. 2017). Sequences were grouped into amplicon sequence variants (ASVs). The chimeras were filtered by VSEARCH method. Taxonomy was annotated to each ASV using the UNITE database (2020-02-20). Singletons were filtered for downstream analysis. A subset of 40,000 sequences per sample was chosen to compare fungal community for all samples.
The differences in fungal community compositions between the two hosts were shown by non-metric multidimensional scaling (NMDS) and analysis of similarity (ANOSIM; permutations = 999) using the vegan package (Version 2.0-2) in R software (v.3.4.3). Identification of gut fungal biomarkers in each species was analyzed by linear discriminant analysis (LDA) effect size (LEfSe; Segata et al. 2011). Indicator analysis was conducted to show the enriched genera in each host. The fungal functional guilds (i.e., functional group) were assigned using the FUNGuild pipeline, and only these guilds with high confidence ranking (i.e., highly probable and probable) were selected for further analysis (Nguyen et al. 2016). One-way ANOVA was conducted to evaluate the differences of fungal alpha-diversity (normal distribution, Kolmogorov–Smirnov test) between the two hosts. The Mann–Whitney–Wilcoxon test was performed to compare the relative abundance of animal pathogen and pathogenic diversity (non-normal distribution) between the two hosts.
Results
Intestinal fungal alpha-diversity
A total of 2,116,221 quality-filtered fungal sequences were obtained across all samples, ranging from 40,701 to 72,417 sequences per sample. A total of 1052 fungal ASVs were found, ranging from 23 to 260 across all samples, 25.3% of which (266) shared in guts of the two species (Additional file 1: Fig. S1). The unique gut fungal ASVs were 591 (56.2%) and 195 (18.5%) for Hooded Crane and Greylag Goose, respectively. Gut fungal alpha-diversity (i.e., ASV richness and Shannon index) was significantly higher in Hooded Crane than Greylag Goose (Fig. 1).
Intestinal fungal community structure
The dominant fungal phyla were Ascomycota (72%), Basidiomycota (22%), Zygomycota (3.6%) and Rozellomycota (1.7%) in guts of the two hosts. The Greylag Goose had significantly higher relative abundance of Ascomycota, lower relative abundance of Basidiomycota, Zygomycota and Rozellomycota relative to Hooded Crane (Additional file 1: Fig. S2). Dramatic shifts of gut fungal community structures were detected between Hooded Crane and Greylag Goose (ANOSIM: P = 0.001; Fig. 2). The FUNGuild analysis showed that the guts of Hooded Crane had higher relative abundance of plant saprotroph than Greylag Goose (Additional file 1: Fig. S3).
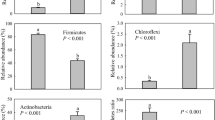
The LEfSe showed that fungi in four phyla (i.e., Basidiomycota, Glomeromycota, Rozellomycota and Zygomycota), six classes (i.e., Dothideomycetes, Leotiomycetes, Sordariomycetes, etc.), and 14 orders (i.e., Helotiales, Pleosporales, Thelebolales, etc.) were enriched in guts of Hooded Crane (Fig. 3). Fungi from one phylum (i.e., Ascomycota), one class (i.e., Eurotiomycetes) and four orders (i.e., Botryosphaeriales, Dothideales, Eurotiales and Wallemiales) were more abundant in guts of Greylag Goose (Fig. 3). Indicator analysis showed that there were 18 (e.g. Acremonium, Phoma, Rhodotorula, etc.) and six (i.e., Aspergillus, Talaromyces, Sagenomella, etc.) indicator genera in Hooded Crane and Greylag Goose, respectively (Additional file 2: Table S1).
Intestinal potential pathogen
The FUNGuild analysis was used to infer the potentially animal fungal pathogens in Hooded Crane and Greylag Goose. There were 42 potentially pathogenic ASVs across all samples, ranging from zero to ten ASVs per sample. The 17% (i.e., 7) of total potentially pathogenic ASV was detected in both host species (Fig. 4a). The guts of Hooded Crane (i.e., 22) had more unique pathogenic ASV than Greylag Goose (i.e., 13; Fig. 4a). The Mann–Whitney–Wilcoxon test showed that relative abundance of potential pathogen and pathogenic diversity (i.e., ASV richness) were significantly higher in Hooded Crane than Greylag Goose (Fig. 4b, c).
a The animal pathogenic ASV overlapping in guts of Hooded Crane and Greylag Goose; b the relative abundance of animal pathogen; and c animal pathogenic diversity (i.e., pathogenic ASV). Letters represent significant differences from the Mann–Whitney–Wilcoxon test (P < 0.05). HC, Hooded Crane; GG, Greylag Goose; ASV, amplicon sequence variant
Discussion
In this study, divergence in the gut fungal community composition and alpha diversity was found between wintering Hooded Crane and Domestic Goose (Figs. 1, 2, 3). Previous studies have shown that diet was the primary driver inducing shifts in microbial community between hosts (Bolnick et al. 2014; Palamidi and Mountzouris 2018). Hooded Crane mainly ate wild Vallisneria natans and Potamogeton malaianus, while artificial hurl food (i.e., paddy) was the main food resource for Domestic Goose (Zheng et al. 2015; Fu et al. 2020). Thus, the dramatic variations in intestinal fungal community might be induced by different diets between the two hosts (Stanley et al. 2012).
The Hooded Crane had higher gut fungal diversity than Greylag Goose (Fig. 1). Recent studies have demonstrated that the external living environment affects animal gut microbial diversity (Rothschild et al. 2018; Perofsky et al. 2019; Schmidt et al. 2019). As Hooded Crane is a kind of migratory bird, which flies long distance and lives at various habitats. However, the living condition and food resources of domestic Greylag Goose were relatively stable. Thus, the more input of environmental microbiota led to higher gut fungal diversity in Hooded Crane than Greylag Goose. A previous study verified that gut microbial diversity was a crucial factor affecting host’s digestion and glycemic control (Liu et al. 2020). The wild food resources exhibit lower nutrient contents relative to paddy. Therefore, the higher intestinal fungal diversity might benefit wintering Hooded Cranes to efficiently acquire nutrients from indigestible food (Cantarel et al. 2012; Li et al. 2019).
Furthermore, the higher relative abundance of plant saprotroph was detected in Hooded Crane than Greylag Goose (Additional file 1: Fig. S3). The roots and leaves of certain plants were eaten by Hooded Crane during wintering period (Zheng et al. 2015), thus higher abundance of plant saprotroph might be associated with faster materials conversion and higher nutrient absorption for Hooded Crane. The Acremonium and Rhodotorula were indicator genera in guts of Hooded Crane (Additional file 2: Table S1). Previous studies have demonstrated that the two genera might increase host’s digestion and immunity (Li et al. 2018; Chen et al. 2020). The wintering Hooded Crane is under great pressure with food shortage and pathogen invasion (Xiang et al. 2019). Together all, the results suggested that Hooded Crane might depend more on their intestinal microbiota to acquire nutrients and enhance immunity.
In this study, Hooded Crane carried more gut animal pathogenic abundance and diversity than Greylag Goose (Fig. 4), suggesting that wild migratory birds might suffer various pathogens under harsh living conditions. Hooded Cranes are the threatened species, so more work should be done to protect them (Xiang et al. 2019). The 17% of total potentially pathogenic ASVs were detected in both hosts (Fig. 4), suggesting that there might be a little bit of cross infection of fungal pathogens between Hooded Crane and Greylag Goose. As anthropic activities induced the degradation of the lake, the serious reduction in food availability for wintering wild birds happened in recent years (Yang et al. 2015). The wintering birds often foraged together with poultry in paddy fields (Fu et al. 2020). Thus, we hypothesized that the cross-infection of pathogens between Hooded Crane and Greylag Goose might occur when they flocked together for foraging.
There were 42 fungal pathogenic ASVs in Hooded Crane and Greylag Goose (Fig. 4). They could propagate their gut pathogens and increase risk of diseases in other sympatric animals (Ekong et al. 2018; Xiang et al. 2019). Local residents have a lot of contact with domestic poultry, thus those pathogens in guts of Greylag Goose could propagate to human beings. Furthermore, we found two genera with high relative abundance (i.e., Aspergillus and Talaromyces) in guts of Greylag Goose (Additional file 2: Table S1). The Aspergillus could produce ochratoxin (i.e., cancerogen; Cafsi et al. 2020) and the Talaromyces induces enterobrosis (Zhao et al. 2020) for human beings. Thus, the intestinal pathogens in poultry might affect the health of human beings.
Conclusions
The results demonstrated that gut fungal community composition and diversity showed significant difference between wintering Hooded Crane and domestic Greylag Goose. The Hooded Crane might depend more on their gut fungal community to acquire nutrients from indigestible food resources. The Hooded Crane lives at various habitats under harsh living conditions, leading to higher gut fungal pathogenic diversity. Our study also implied that more research should focus on intestinal pathogens in wild birds and domestic poultry, as these pathogens might cause disease in other animals, even human beings. This work helps our further understanding in intestinal microbial community of migratory birds and domestic poultry. However, there were certain limitations. We did not clearly verify the cross infection of gut pathogens between wild birds and domestic poultry along wintering timescale. In addition, the gut fungal communities of Hooded Crane and domestic poultry were studied within one wintering region rather than across multiple regions. The limitations should be clarified in future studies.
Availability of data and materials
The raw data were submitted to the Sequence Read Archive (SRA) under the accession number SRP276474.
References
Alm EW, Daniels-Witt QR, Learman DR, Ryu H, Jordan DW, Gehring TM, et al. Potential for gulls to transport bacteria from human waste sites to beaches. Sci Total Environ. 2018;615:123–30.
Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296–302.
Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Xu ZZ, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191-16.
Ayayee PA, Larsen T, Rosa C, Felton GW, Ferry JG, Hoover K. Essential amino acid supplementation by gut microbes of a wood-feeding cerambycid. Environ Entomol. 2016;45:66–73.
Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:1091.
Cafsi IE, Bjeoui S, Rabeh I, Nechi S. Effects of ochratoxin A on membrane phospholipids of the intestine of broiler chickens, practical consequences. Animal. 2020;14:933–41.
Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE. 2012;7:e28742.
Chen M, Chen XQ, Tian LX, Liu YJ, Niu J. Enhanced intestinal health, immune responses and ammonia resistance in Pacific white shrimp (Litopenaeus vannamei) fed dietary hydrolyzed yeast (Rhodotorula mucilaginosa) and Bacillus licheniformi. Aquacult Rep. 2020;17:100385.
Chung HC, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93.
Dong Y, Xiang X, Zhao G, Song Y, Zhou L. Variations in gut bacterial communities of Hooded Crane (Grus monacha) over spatial-temporal scales. PeerJ. 2019;7:e7045.
Eckburg P, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Ekong PS, Fountain-Jones NM, Alkhamis MA. Spatiotemporal evolutionary epidemiology of H5N1 highly pathogenic avian influenza in West Africa and Nigeria, 2006–2015. Transbound Emerg Dis. 2018;65:70–82.
Fox AD, Cao L, Zhang Y, Barter M, Zhao MJ, Meng FJ, et al. Declines in the tuber-feeding waterbird guild at Shengjin Lake National Nature Reserve, China—a barometer of submerged macrophyte collapse. Aquat Conserv. 2011;21:82–91.
Fu R, Xiang XJ, Dong YQ, Cheng L, Zhou LZ. Comparing the intestinal bacterial communies of sympatric wintering Hooded Crane (Grus monacha) and Domestic Goose (Anser anser domesticus). Avian Res. 2020;11:13.
Geib SM, Filley TR, Hatcher PG, Hoover K, Carlson JE, Jimenez-Gasco MDM, et al. Lignin degradation in wood-feeding insects. Proc Natl Acad Sci USA. 2008;105:12932–7.
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:1657–68.
Herr JR, Scully ED, Geib SM, Hoover K, Carlson JE, Geiser DM. Genome sequence of Fusarium isolate MYA-4552 from the midgut of Anoplophora glabripennis, an invasive, wood-boring beetle. Genome Announc. 2016;4:e00544-e616.
Li Y, Sun YK, Li X, Zhang GN, Xin HS, Xu HJ, et al. Effects of Acremonium terricola culture on performance, milk composition, rumen fermentation and immune functions in dairy cows. Anim Feed Sci Technol. 2018;240:40–51.
Li Y, Zhang K, Liu Y, Li K, Hu D, Wronski T. Community composition and diversity of intestinal microbiota in captive and reintroduced Przewalski’s horse (Equus ferus przewalskii). Front Microbiol. 2019;10:1821.
Liu Y, Wang Y, Ni Y, Cheung CKY, Lam KSL, Wang Y, et al. Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab. 2020;31:77–91.
Mason CJ, Campbell AM, Scully ED, Hoover K. Bacterial and fungal midgut community dynamics and transfer between mother and brood in the Asian Longhorned Beetle (Anoplophora glabripennis), an invasive xylophage. Microb Ecol. 2019;77:230–42.
Nguyen NH, Song ZW, Bates ST, Branco S, Tedersoo L, Menke J, et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–8.
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7.
Palamidi I, Mountzouris KC. Diet supplementation with an organic acids-based formulation affects gut microbiota and expression of gut barrier genes in broilers. Anim Nutr. 2018;4:367–77.
Perofsky AC, Lewis RJ, Meyers LA. Terrestriality and bacterial transfer: a comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J. 2019;13:50–63.
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5.
Schmidt E, Mykytczuk N, Schulte-Hostedde AI. Effects of the captive and wild environment on diversity of the gut microbiome of deer mice (Peromyscus maniculatus). ISME J. 2019;13:1293–305.
Scully ED, Hoover K, Carlson J, Tien M, Geib SM. Proteomic analysis of Fusarium solani isolated from the Asian longhorned beetle Anoplophora glabripennis. PLoS ONE. 2012;7:e32990.
Segata N, Izard J, Walron L, Gevers D, Miropolsky L, Garrett W, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA. 2010;107:20051–6.
Singh P, Karmi A, Devendra K, Waldroup PW, Cho KK, Kwon YM. Influence of penicillin on microbial diversity of the cecal microbiota in broiler chickens. Poult Sci. 2013;92:272–6.
Stanley D, Denman SE, Hughes RJ, Geier MS, Crowley TM, Chen H, et al. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl Microbiol Biot. 2012;96:1361–9.
Tanahashi M, Kubota K, Matsushita N, Togashi K. Discovery of mycangia and the associated xylose-fermenting yeasts in stag beetles (Coleoptera: Lucanidae). Naturwissenschaften. 2010;97:311–7.
Tanahashi M, Kim JK, Watanabe K, Fukatsu T, Kubota K. Specificity and genetic diversity of xylose-fermenting Scheffersomyces yeasts associated with small blue stag beetles of the genus Platycerus in East Asia. Mycologia. 2017;109:630–42.
Xiang XJ, Zhang FL, Fu R, Yan SF, Zhou LZ. Significant differences in bacterial and potentially pathogenic communities between sympatric Hooded Crane and Greater White-fronted Goose. Front Microbiol. 2019;10:163.
Xu LT, Lu M, Xu DD, Chen L, Sun JH. Sexual variation of bacterial microbiota of Dendroctonus valens guts and frass in relation to verbenone production. J Insect Physiol. 2016;5:110–7.
Yang L, Zhou L, Song Y. The effects of food abundance and disturbance on foraging flock patterns of the wintering Hooded Crane (Grus monacha). Avian Res. 2015;6:15.
Zhang FL, Xiang XJ, Dong YQ, Yan SF, Song YW, Zhou LZ. Significant differences in the gut bacterial communities of Hooded Crane (Grus monacha) in different seasons at a stopover site on the flyway. Animals. 2020;10:701.
Zhao YK, Liu JY, Liu JH, Lu S, Wu HH, Luo DQ. Recurrent Talaromyces marneffei infection presenting with intestinal obstruction in a patient with systemic lupus erythematosus. Mycopathologia. 2020;185:717–26.
Zheng M, Zhou L, Zhao N, Xu W. Effects of variation in food resources on foraging habitat use by wintering Hooded Cranes (Grus monacha). Avian Res. 2015;6:11.
Acknowledgements
We thank Mr. Wei Wang from Anhui University for assistance in sample collection, and Prof. Binghua Sun from Anhui University for assistance with manuscript revision.
Funding
This work was supported by the National Natural Science Foundation (31801989), the Natural Science Foundation of Education Committee (KJ2018A0001) and the Scientific Research Foundation (J01006160, J05011710, S020118002/101).
Author information
Authors and Affiliations
Contributions
XX and ZF designed the experiment; JL, YZ and ZN collected the samples; XX and JL performed the experiments; XX, YZ and ZF performed data analysis; XX and ZF wrote the manuscript; JL, YZ and ZN revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Non-invasive sample collection without hunting of experimental animals was used in this study. Permission was obtained from the Shengjin Lake National Nature Reserve.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1: Figure S1.
Venn diagram showing the co-occurrence of the ASVs between the two species. Figure S2. Relative abundances of the dominant fungal phyla. Figure S3. The relative abundance of plant saprotroph in guts of Hooded Crane (HC) and Greylag Goose (GG).
Additional file 2: Table S1.
Indicator analysis was conducted to show indicator genera with relative abundance > 0.1% of each species.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xiang, X., Jin, L., Yang, Z. et al. Dramatic shifts in intestinal fungal community between wintering Hooded Crane and Domestic Goose. Avian Res 12, 1 (2021). https://doi.org/10.1186/s40657-020-00238-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40657-020-00238-1