Abstract
Background
Carbapenem-resistant Acinetobacter baumannii has recently been defined by the World Health Organization as a critical pathogen. The aim of this study was to compare clonal diversity and carbapenemase-encoding genes of A. baumannii isolates collected from colonized or infected patients and hospital environment in two intensive care units (ICUs) in Morocco.
Methods
The patient and environmental sampling was carried out in the medical and surgical ICUs of Mohammed V Military teaching hospital from March to August 2015. All A. baumannii isolates recovered from clinical and environmental samples, were identified using routine microbiological techniques and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Antimicrobial susceptibility testing was performed using disc diffusion method. The carbapenemase-encoding genes were screened for by PCR. Clonal relatedness was analyzed by digestion of the DNA with low frequency restriction enzymes and pulsed field gel electrophoresis (PFGE) and the multi locus sequence typing (MLST) was performed on two selected isolates from two major pulsotypes.
Results
A total of 83 multidrug-resistant A. baumannii isolates were collected: 47 clinical isolates and 36 environmental isolates. All isolates were positive for the bla OXA51-like and bla OXA23-like genes. The coexistence of bla NDM-1 /bla OXA-23-like and bla OXA 24-like /bla OXA-23-like were detected in 27 (32.5%) and 2 (2.4%) of A. baumannii isolates, respectively. The environmental samples and the fecally-colonized patients were significantly identified (p < 0.05) as the most common sites of isolation of NDM-1-harboring isolates. PFGE grouped all isolates into 9 distinct clusters with two major groups (0007 and 0008) containing up to 59% of the isolates. The pulsotype 0008 corresponds to sequence type (ST) 195 while pulsotype 0007 corresponds to ST 1089.The genetic similarity between the clinical and environmental isolates was observed in 80/83 = 96.4% of all isolates, belonging to 7 pulsotypes.
Conclusion
This study shows that the clonal spread of environmental A. baumannii isolates is related to that of clinical isolates recovered from colonized or infected patients, being both associated with a high prevalence of the bla OXA23-like and bla NDM-1 genes. These findings emphasize the need for prioritizing the bio-cleaning of the hospital environment to control and prevent the dissemination of A. baumannii clonal lineages.
Similar content being viewed by others
Background
Multidrug-resistant (MDR) Acinetobacter baumannii is recognized to be responsible for nosocomial outbreaks in severely ill patients and it is predominantly isolated in intensive care units (ICUs) around the world [1]. This microorganism colonizes certain areas of the body such as the skin, the oropharynx and the gastrointestinal tract [1]. The prevalence of digestive tract colonization varies from 8.3 to 41% in ICU patients [2, 3] but this pathogen is also the causative agent of serious infections including pneumonia, septicemia, urinary tract infection, wound infection and meningitis with mortality rates varying from 7.8 to 75% [1]. Risk factors for Acinetobacter colonization and infection are linked to the presence of underlying disease, long-term hospitalization, ICU stay, administration of broad spectrum antibiotics and invasive procedures such as mechanical ventilation or catheters [1, 4]. This bacterium displays an outstanding ability to survive in the environment, with some studies reporting up to 48% of environmental samples being contaminated with Acinetobacter [5, 6]. Environmental sites most likely to be contaminated include bed sheets, bed railings, touch pads of ventilator equipment, trolleys, surfaces of respiratory monitors as well as the hands and uniforms of healthcare workers [5,6,7].
A. baumannii has also the capacity to develop resistance to multiple antibiotics, which limits the therapeutic options to treat these infections [1, 4]. A recent Moroccan study showed that the resistance rate of Acinetobacter isolates to ciprofloxacin, imipenem, amikacin netilmicin, and colistin was 87%, 86%, 52%, 33% and 1.7%,respectively [8]. Resistance to carbapenems among A. baumannii isolates all over the world is mostly linked to the carriage of the bla OXA-23-like, bla OXA-24-like, and bla OXA-58-like genes, encoding carbapenem hydrolyzing class D β-lactamases (OXA-type), but also to the recent dissemination of the bla NDM gene, encoding a class B metallo-β-lactamase [9,10,11,12,13,14]. Since 2010, NDM-producing A. baumannii isolates have been found in different countries including Kenya, Ethiopia, Algeria, Egypt, Germany, France, Spain, Turkey, India,Vietnam, China and Nepal [14, 15].
Overall, the clonal dissemination of carbapenem-resistant A. baumannii isolates has been documented in different countries [6, 7, 9,10,11] but only a few studies have focused on the clonal relationships between clinical and environmental isolates [5, 16, 17].
To our knowledge, there are no previous studies regarding the prevalence of carbapenemase encoding genes or the clonal diversity of A. baumannii isolates in Morocco.
The objective of this study was to characterize the carbapenemase-encoding genes and molecular diversity of clinical and environmental carbapenem-resistant A. baumannii isolates recovered from two ICUs of a Moroccan hospital.
Methods
This study was carried out in the clinical bacteriology laboratory of Mohammed V Military teaching hospital in collaboration with Barcelona Institute for Global Health (IS global)-Hospital Clínic, Universitat de Barcelona.
Sampling strategies
The patient and environmental sampling was carried out from March to August 2015 in the medical and surgical ICUs of Mohammed V Military teaching hospital, a teaching hospital with 700-beds, located in Rabat in the Kingdom of Morocco, and which contains 2 ICUs (medical and surgical) with 10 beds each, a center for burns, surgical and medical units, and laboratory and imagery departments.
The clinical isolates were recovered from the mouth, the anal margin and the groin for colonized patients and from the respiratory tract and blood cultures for infected patients. The criteria of colonization or infection were assessed according to the Centers for Disease Control and Prevention guidelines [18]. Screening samples were collected at the time of ICU admission and weekly during hospitalization. Collected clinical data included demographic characteristics, hospital wards, underlying diseases, invasive procedures, specimen types, antibiotic use, ICU length of stay and clinical outcome.
Environmental samples were collected from the patients’ rooms. At each site, an area of 10 cm2 was sampled using a sterile swab moistened with physiological saline [19, 20]. The sampled sites were: floors, bed sheets, medical ventilators, pillows, monitors, patient trolleys and intravenous solution stand.
All swabs were then immersed in brain heart infusion broth, incubated overnight at 37 °C and further subcultured on bromocresol purple lactose agar for the isolation of Acinetobacter.
All Acinetobacter spp. isolates were identified using routine microbiological techniques (direct examination, biochemical test of orientation, API20NE) and species identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) [21].
Antibiotic susceptibility testing
Antimicrobial susceptibility testing was performed by the disc diffusion method on Mueller-Hinton agar plates in accordance with the French Society of Microbiology in their 2015 recommendations guidelines. MDR A. baumannii isolates were defined as resistant to three or more classes of antibiotics represented by piperacillin/tazobactam,ceftazidime, imipenem, ciprofloxacin, aminoglycosides and colistin [22].
PCR assays for detection of carbapenemase-encoding genes
DNA extractions from overnight cultures were performed using PureLink® Genomic DNA Kit (Invitrogen, Carlsbad, USA) and DNA IQ™ System (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. PCR analysis was carried out as described previously [23, 24] using the thermocycler (Biometra, Göttingen, Germany) with the primers listed in Table 1.
Multiplex PCR assays were used to detect four carbapenemase-encoding genes (bla OXA-51-like, blaOXA-23-like, blaOXA-24-like and blaOXA-58-like). PCR amplifications for blaOXA genes were performed in a final volume of 50 μl, reaction mixtures contained 5 μl of 10× PCR buffer, 25 mmol/μL of MgCI2, 2.5 Mm of deoxynucleoside triphosphates (dNTPs), 0.5 μl of each primer, 1 U Taq DNA polymerase (New England BioLabs Inc., Beverly, MA, USA) and 3 μl of DNA template. The amplification conditions were initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 25 s, 52 °C for 40s and 72 °C for 50s, with final extension for 6 min at 72 °C.
Uniplex PCR was used for detection of NDM-1 gene. PCR reaction for bla NDM-1 gene was carried out by adding 0.5 μl of each primer, 5 μl of 10Xbuffer, 3 μl of MgCl2, 1.25 μl of dNTP’s, and 0.25 μl of Taq polymerase in a final volume of 50 μl. PCR conditions were as follows: initial denaturation at 94 °C for 10 min, followed by 32 cycles consisting of denaturation at 94 °C for 30 s, 40 s annealing at 57 °C, 50 s extension at 72 °C, followed by a final extension step at 72 °C for 5 min.
Hyperladder 100 bp (Bioline, London, UK) was used as a molecular weight marker. PCR amplification products were analyzed by gel electrophoresis in a 1.5% w/v Agarose gel stained with SYBR safe.
Molecular typing using pulsed-field gel electrophoresis
The clonal relationship of all isolates was analyzed by PFGE as previously described with minor modifications [25]. An overnight culture on blood agar was suspended in 120 μl of cell suspension buffer (100 mM Tris-HCl, 100 mM EDTA, pH 8.0) and then, the bacterial suspension was mixed with an equal volume of 2% InCert™ Agarose (Lonza,Rockland,ME,USA) and dispensed in a plug mould. Genomic DNA in agarose plugs was lysed in the cell lysis solution (50 Mm Tris-HCl, 1% sarcosil, 100 μg/ml proteinase K), washed and digested with ApaI (New England BioLabs Inc., Beverly, MA, USA). Electrophoresis was performed in 1% InCert™ Agarose (Lonza, Rockland,ME,USA) and 0.5X TBE Buffer (PH 8.0) containing 0.02 g of thiourea using either a CHEF-DR III system (Bio-Rad Laboratories) or a CHEF-Mapper TM apparatus (Bio-Rad Laboratories) at 6 V/cm2 with switch times ranging from 5 s to 35 s at an angle of 120°, at temperature of 14 °C, for 20 h.
A standard molecular weight Lambda DNA ladder (Bio-Rad Laboratories) was included at least twice per gel to allow normalization of all fingerprints. The InfoQuest™FP v.4.5 software (Bio-Rad Laboratories) was used for dendrogram construction by the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method, based on Dice’s similarity coefficient. Isolates were considered to belong to the same PFGE cluster (pulsotype) if their Dice similarity index was ≥ 85% [26].
On the basis of the number of isolates, PFGE pulsotypes were divided into major pulsotypes (more than ten isolates/PFGE types), intermediate pulsotypes (five to nine isolates/PFGE types) and minor pulsotypes (less than five isolates/PFGE types) (Table 2).
Multi locus sequence typing (MLST)
The whole-genome sequencing was performed on the extracted genomic DNA of the two selected strains of major pulsotypes by using the Nextera XT DNA library preparation kit (Illumina), with dual indexing adapters, and sequenced using an Illumina MiSeq sequencer with a 2 × 251-bp paired-end configuration. The Next Generation Sequencing Data (FASTA format) were then used for further MLST analysis, carried out using MLST Oxford scheme (https://pubmlst.org/abaumannii/).
Statistical analysis
Statistical analysis was performed using SPSS Statistics for Windows, version 10.0. The results were expressed as effective and percentages for qualitative variables and as mean (standard deviation) or median (interquartile range: IQR) for quantitative variables. The chi-square and Fisher exact tests were used to compare the qualitative variables. A comparison between the two quantitative variables was performed using the Mann–Whitney U test for non-normal distributed variables, whereas Student’s t-test was used for normally distributed variables. P values less than 0.05 were considered significant.
Results
Bacterial isolates and epidemiological data
A total of 83 non-duplicate A. baumannii isolates were collected: 47 clinical isolates from 40 colonized or infected patients and 36 isolates from 72 environmental specimens.
Among 40 patients, 32 (80%) were colonized and 8 (20%) were infected. The epidemiological and clinical characteristics of patients are shown in Table 3. The average age was 54.38 ± 15.22 years and 30 (75%) were males representing a sex ratio M/F of 3:1. The crude mortality rate was 54.2%. The crude mortality rate was 43.8% in colonized patients and 87.5% in infected ones (p = 0.031). Clinical isolates were recovered from 39 screening samples including anal margin (22/47 = 46.8%), mouth (10/47 = 21.3%), groin (7/47 = 14.9%) and from 8 diagnosis samples composed of blood culture (3/47 = 6.4%), protected distal sampling (3/47 = 6.4%) and bronchial aspiration (2/47 = 4.2%).
Of the 72 environmental samples, 36 (50%) yielded A. baumannii isolates. Surgical ICU samples were more contaminated (22/31 = 71%) than those from the medical ICU (14/41 = 34.1%) (p = 0.004). The environmental A. baumannii isolates were obtained from bed sheets (14/36 = 38.9%), floors (13/36 = 36.1%), medical ventilators (4/36 = 11.1%), patient trolleys (2/36 = 5.5%), pillows (1/36 = 2.8%), monitors (1/36 = 2.8%), and intravenous solution stands (1/36 = 2.8%).
Antimicrobial susceptibility profile
All isolates were MDR. The difference in resistance rates between the clinical isolates and the environmental ones was not statistically significant except for gentamicin (85.1% vs 100% respectively, p = 0.015).
Distribution of carbapenemase genes
The intrinsic chromosomally encoded bla OXA-51 like gene characteristic of A. baumannii was detected in all isolates. All isolates were positive for bla OXA-23-like.The coexistence of bla NDM-1 with bla OXA-23-like and bla OXA 24-like with bla OXA-23 was detected in 27 (32.5%) and 2 (2.4%) of A. baumannii isolates, respectively. The bla OXA-58-like gene was not detected. The percentage of NDM-1 carriage was significantly higher in environmental isolates than in clinical ones (18/36 = 50% vs 9/47 = 19.1%, p = 0.004). NDM-1 producing clinical isolates were recovered from: anal margin (88.9%) and mouth (11.1%). The NDM-1 harboring isolates were significantly (p = 0.006) more frequently isolated from anal margin samples than from other clinical specimens. The distribution of NDM-1 positive isolates among hospital environment samples is as follows: bed sheets (38.9%), floor (16.7%), medical ventilators (16.7%), intravenous solution stands (5.5%), monitors (5.5%), patient trolleys (5.5%) and pillows (5.5%). No significant differences (p = 0.432) in the number of NDM-1 positive isolates were found between different environmental sampling sites.
PFGE and MLST analyses
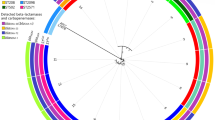
The isolates were classified into 9 PFGE pulsotypes (0001–0009) with two major pulsotypes (0008, 0007), containing up to 59% of all isolates. The strains of pulsotype 0008 belonged to sequence type (ST) 195 while those of pulsotype 0007 belonged to ST 1089. Clinical isolates were found in all 9 different pulsotypes while environmental isolates were only present in 7 pulsotypes, as they were missing from pulsotypes 0004 and 0006 which included just 2 and 1 isolates, respectively (Table 2) (Fig. 1) (Additional file 1) .
Discussion
In the present study we have analyzed the clonal relatedness and resistance characteristics of A. baumannii isolates recovered. The clinical isolates were commonly isolated from the anal margin (46.8%) followed by the mouth (24%). The digestive tract of patients hospitalized in the ICU, has been identified as an important site for Acinetobacter colonization which can lead to severe infections, with a prevalence of 8.3% in Saudi Arabia [2] and 41% in Spain [3]. A study conducted in Spain by Corbella et al. showed that MDR A. baumannii infections occurred more frequently in patients with fecal colonization than in those without fecal colonization [3]. The crude mortality found in this study (50%) was comparable to that reported in colonized or infected patients in Italy (58%) [27] but significantly higher than that of Spanish hospitals (18.9%) [4].The mortality rate of patients depends on clinical significance of A. baumannii. In the current study, the mortality rate was significantly higher in infected patients than in colonized ones (87.5% vs 43.8%, p = 0. 031). This result is similar to that of Rodríguez-Baño J et al. in Spain who showed that crude mortality was higher in infected (27%) than in colonized patients (10%) [4].
Our findings show a higher environmental contamination around infected or colonized patients in our ICU. The presence of A. baumannii among environmental specimens (50%) was higher than that observed in similar studies: 7.7% in Algeria [7], 9.9% in the United States of America [19], 13.1% in China [16] and between 2% and 18% in two different studies in Turkey [5, 6]. This can be attributed to the lack of hospital decontamination procedures and hand hygiene in our region. This study also shows that the surgical ICU samples were more contaminated than those of medical ICU (71% vs 37.2%, p = 0.004) and the sites frequently touched by both the health-care workers and patients were the most contaminated as the majority of environmental isolates were recovered from floors (42.1%) followed by bed sheets (34.2%) and medical ventilators (10.5%). These findings are in agreement with that of other researchers who reported that this pathogen was isolated from near-patient surfaces, medical equipment, airborne samples and healthcare workers’ hands [5,6,7, 19].
In the current study, all isolates were resistant to imipinem. Carbapenem-resistance among A. baumannii isolates has shown a steady increase in our region since 2001, when it was reported around 23.6% [28] and then increased to 76.19% in 2012–2014 [8]. This can be explained by the excessive use of inadequate empirical antimicrobial treatment including carbapenems, poor infection control practices, poor antimicrobial stewardship governance and widespread dissemination of carbapenem-resistant strains in the community.
In the present study, carbapenem-resistance was mainly attributed to the carriage of the bla OXA-23-like gene that was present in all isolates (100%).This prevalence is similar to that reported in Turkey (100%) [11] and Egypt (100%) [12] but higher than that observed in Brazil (95.4%) [29], Asian pacific countries (95%) [9], France (82%) [30], South Africa (77%) [31] and Italy (71.7%) [10]. During the past decades, outbreak or sporadic A. baumannii clones producing OXA-23 have disseminated around the world [32] but such dissemination has been particularly relevant among Mediterranean countries, where the bla OXA-23-like gene has replaced previously predominant blaOXA genes such as bla OXA-24-like and bla OXA-58-like [33]. In our study, only 2 clinical isolates also harbored the bla OXA-24-like gene (4.25%) but this gene was not detected among the environmental isolates and all isolates were negative for the bla OXA-58-like gene. The high prevalence of bla OXA-23-like gene is probably associated with horizontal gene transfer by mobile genetic elements such as plasmids, transposable elements and integron systems. It has been reported that the spread of bla OXA-23-like genes is associated with the Tn2006, Tn2007, Tn2008, and Tn2009 transposons, which can be further located on the chromosome or on conjugative plasmids [32, 34, 35].
Likewise, 32.5% of all isolates also presented the bla NDM-1 gene and this is the first time that NDM-producing A. baumannii isolates are reported in Morocco. Our results indicate that NDM-1-producing A. baumannii isolates are widely circulating in the hospital environment and they were found in all environmental sampling sites. Moreover, the NDM-1-producing A. baumannii isolates were more frequently recovered (p = 0.004) from environmental isolates (50%) than from clinical isolates (19.1%).The environmental NDM-1 producing A. baumannii isolates have also been reported in Algeria [7] and in China [36]. The high rate of environmental bla NDM-1 contamination is alarming as the hospital environment may become a potential reservoir for A. baumannii isolates carrying NDM-1 which could result in transfer the bla NDM-1 gene to other bacterial species.
Among clinical isolates, the anal margin samples were identified (p = 0.006) as the most common sites of isolation of NDM-1-producing A. baumannii isolates. These findings also highlight the role played by fecally-colonized patients as reservoirs for carbapenem-resistant nosocomial A. baumannii isolates.
In the current study, the genetic similarity between clinical and environmental isolates was observed in (80/83 = 96.4%) of all isolates, classified into 7 pulsotypes (0001, 0002, 0003, 0005, 0007, 0008 and 0009). These results suggest a dynamic exchange of A. baumannii isolates between patients and their environmental surroundings. These pathogens can be transmitted from patient-to-patient, patient to a health care worker, patient to environment and vice versa. In our study, three clinical isolates belonging to the pulsotypes (0004 and 0006) which were not detected in environmental isolates, may be exclusively transmitted through direct contact between an infected or colonized patient and another person or they may come from other environmental reservoirs which are either rare or not identified in our study.
Our results also show that the most frequent pulsotype was PFGE type 0008 (30/83 = 36.1%). Among the two major pulsotypes, clinical isolates were predominant within pulsotype 0007 (16/47 = 34%) while environmental isolates were a majority within pulsotype 0008 (16/36 = 44.4%). Overall, however, both major pulsotypes were closely related (Dice similarity >82%) and contained up to 59% of all isolates, they were found in all sampling sites and it is clear that they have become endemic in this particular setting. The PFGE cluster 0008 corresponds to ST195 (Oxford MLST) which has been previously reported from Asian countries, European nations and Egypt [37,38,39] while the strains from pulsotype 0007 were assigned to ST 1089(Oxford MLST) which is very rare ST and according to “the profile history for A. baumannii MLST (Oxford) database”, the ST 1089 has been found for the first time in India in 2015.
Conclusion
This study shows that the clonal spread of environmental A. baumannii isolates is related to that of clinical isolates recovered from colonized or infected patients. Our results have also shown that OXA-23 is the most common carbapenemase among A. baumannii isolates in our hospital but the prevalence of isolates producing both OXA-23 and NDM-1 is also alarming.
Effective control measures are urgently needed to prevent the transmission of endemic lineages of MDR A. baumannii and they should take into account the decontamination of the patients’ environmental surroundings.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- dNTPs:
-
deoxynucleoside triphosphates
- ICU:
-
Intensive care unit
- IQR:
-
Interquartile range
- MALDI-TOF-MS:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MDR:
-
Multidrug-resistant
- MLST:
-
Multi locus sequence typing
- PCR:
-
Polymerase chain reaction
- PFGE:
-
Pulsed-field gel electrophoresis
- ST:
-
Sequence type
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic Mean
References
Manchanda V, Sanchaita S, Singh N. Multidrug Resistant Acinetobacter. J Glob Infect Dis. 2010;2(3):291–304.
Aljindan R, Bukharie H, Alomar A, Abdalhamid B. Prevalence of digestive tract colonization of Carbapenem-Resistant acinetobacter baumannii in hospitals in Saudi Arabia. J Med Microbiol. 2015;64(Pt 4):400–6.
Corbella X, Pujol M, Ayats J, Sendra M, Ardanuy C, Domínguez MA, Liñares J, Ariza J, Gudiol F. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin Infect Dis. 1996;23(2):329–34.
Rodríguez-Baño J, Cisneros JM, Fernández-Cuenca F, Ribera A, Vila J, Pascual A, Martínez-Martínez L, Bou G, Pachón J, Grupo de Estudio de Infección Hospitalaria (GEIH). Clinical Features and Epidemiology of Acinetobacter baumannii Colonization and Infection in Spanish Hospitals. Infect Control Hosp Epidemiol. 2004;25(10):819–24.
Kirkgoz E, Zer Y. Clonal comparison of Acinetobacter strains isolated from intensive care patients and the intensive care unit environment. Turk J Med Sci. 2014;44(4):643–8.
Ertürk A, Çiçek AÇ, Gümüş A, Cüre E, Şen A, Kurt A, Karagöz A, Aydoğan N, Sandallı C, Durmaz R. Molecular characterisation and control of Acinetobacter baumannii isolates resistant to multi-drugs emerging in inter-intensive care units. Ann Clin Microbiol Antimicrob. 2014;13:36.
Zenati K, Touati A, Bakour S, Sahli F, Rolain JM. Characterization of NDM-1- and OXA-23-producing Acinetobacter baumannii isolates from inanimate surfaces in a hospital environment in Algeria. J Hosp Infect. 2016;92(1):19–26.
Uwingabiye J, Frikh M, Lemnouer A, Bssaibis F, Belefquih B, Maleb A, Dahraoui S, Belyamani L, Bait A, Haimeur C, Louzi L, Ibrahimi A, Elouennass M. Acinetobacter infections prevalence and frequency of the antibiotics resistance: comparative study of intensive care units versus other hospital units. Pan Afr Med J. 2016;23:191.
Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23, −24/40 and −58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63(1):55–9.
D'Arezzo S, Principe L, Capone A, Petrosillo N, Petrucca A, Visca P. Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J Antimicrob Chemother. 2011;66(1):54–61.
Aksoy MD, Çavuşlu Ş, Tuğrul HM. Investigation of metallo beta lactamases and oxacilinases in carbapenem resistant Acinetobacter baumannii strains isolated from inpatients. Balkan Med J. 2015;32(1):79–83.
Fouad M, Attia AS, Tawakkol WM, Hashem AM. Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int J Infect Dis. 2013;17(12):e1252–4.
Mesli E, Berrazeg M, Drissi M, Bekkhoucha SN, Rolain JM. Prevalence of carbapenemase-encoding genes including New Delhi metallo-β-lactamase in Acinetobacter species. Algeria Int J Infect Dis. 2013;17(9):e739–43.
Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, Rolain J. New Delhi Metallo-beta-lactamase around the world : An eReview using Google Maps. Euro Surveill. 2014;19(20):20809.
Wei WJ, Yang HF, Ye Y, Li JB. New Delhi Metallo-β-Lactamase-Mediated Carbapenem Resistance: Origin, Diagnosis, Treatment and Public Health Concern. Chin Med J. 2015;128(14):1969–76.
Ying C, Li Y, Wang Y, Zheng B, Yang C. Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J Antibiot (Tokyo). 2015;68(9):562–7.
Tjoa E, Moehario LH, Rukmana A, Rohsiswatmo R. Acinetobacter baumannii: Role in blood stream infection in Neonatal Unit, Dr. Cipto Mangunkusumo Hospital, Jakarta, Indonesia. Int. J Microbiol. 2013;2013:180763.
Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36(5):309–32.
Thom KA, Johnson JK, Lee MS, Harris AD. Environmental contamination because of multidrug-resistant Acinetobacter baumannii surrounding colonized or infected patients. Am J Infect Control. 2011;39(9):711–5.
Sehulster L, Chinn RY, CDC; HICPAC. Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). MMWR Recomm Rep. 2003;52(RR-10):1–42.
Marí-Almirall M, Cosgaya C, Higgins PG, Van Assche A, Telli M, Huys G, Lievens B, Seifert H, Dijkshoorn L, Roca I, Vila J. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin Microbiol Infect. 2017;23(3):210.e1–9.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–3.
Diene SM, Bruder N, Raoult D, Rolain JM. Real-time PCR assay allows detection of the New Delhi metallo-β-lactamase (NDM-1)-encoding gene in France. Int J Antimicrob Agents. 2011;37(6):544–6.
Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4328–35.
Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, Aktas E, Gursoy NC, Caliskan A. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis. 2009;62(5):372–7.
Zarrilli R, Casillo R, Di Popolo A, Tripodi MF, Bagattini M, Cuccurullo S, Crivaro V, Ragone E, Mattei A, Galdieri N, Triassi M, Utili R. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin Microbiol Infect. 2007;13(5):481–9.
Elouennass M, Bajou T, Lemnouer AH, Foissaud V, Hervé VBA. Acinetobacter baumannii : étude de la sensibilité des souches isolées à l’hôpital militaire d’instruction MohammedV, Rabat, Maroc. Med Mal Infect. 2003;33:361–4.
Corrêa LL, Botelho LA, Barbosa LC, Mattos CS, Carballido JM, de Castro CL, Mondino PJ, de Paula GR, de Mondino SS, de Mendonça-Souza CR. Detection of bla(OXA-23) in Acinetobacter spp. isolated from patients of a university hospital. Braz J Infect Dis. 2012;16(6):521–6.
Jeannot K, Diancourt L, Vaux S, Thouverez M, Ribeiro A, Coignard B, Courvalin P, Brisse S. Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS One. 2014;9(12):e115452.
Lowings M, Ehlers MM, Dreyer AW, Kock MM. High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa - an update. BMC Infect Dis. 2015 Nov 14;15:521.
Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 Carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16(1):35–40.
Djahmi N, Dunyach-Remy C, Pantel A, Dekhil M, Sotto A, Lavigne JP. Epidemiology of Carbapenemase-Producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean Countries. Biomed Res Int. 2014;2014:305784.
Liu LL, Ji SJ, Ruan Z, Fu Y, Fu YQ, Wang YF, Yu YS. Dissemination of blaOXA-23 in Acinetobacter spp. in China: Main Roles of Conjugative Plasmid pAZJ221 and Transposon Tn2009. Antimicrob Agents Chemother. 2015;59(4):1998–2005.
Guerrero-Lozano I, Fernández-Cuenca F, Galán-Sánchez F, Egea P, Rodríguez-Iglesias M, Pascual Á. Description of the OXA-23 β-lactamase gene located within Tn2007 in a clinical isolate of Acinetobacter baumannii from Spain. Microb Drug Resist. 2015;21(2):215–7.
Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X, Shi Y, Hu X, An D, Li Z, Li P, Wang L, Cui J, Wang P, Huang L, Klena JD, Song H. Higher Isolation of NDM-1 Producing Acinetobacter baumannii from the Sewage of the Hospitals in Beijing. PLoS One PLoS One. 2013;8(6):e64857.
Ghaith DM, Zafer MM, Al-Agamy MH, Alyamani EJ, Booq RY, Almoazzamy O. The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann Clin Microbiol Antimicrob. 2017;16(1):34.
Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, Holm A, Jakobsen L, Justesen US. Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother. 2015;70(7):1965–8.
Rieber H, Frontzek A, Pfeifer Y. Molecular Investigation of Carbapenem-Resistant Acinetobacter spp. from Hospitals in North Rhine-Westphalia, Germany. Microb Drug Resist. 2017;23(1):25–31.
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from Mohammed V Military Teaching Hospital and from the Ministry of Higher education of Morocco.
This study was also supported by grant 2014SGR0653 from the Departament de Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya, by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, co-financed by European Regional Development Fund (ERDF) “A Way to Achieve Europe,” the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015).
The research leading to these results has also received funding from the People Programme (Marie Curie Actions) of the European Union’s seventh Framework Programme FP7/2007–2013 under REA grant agreement n° 612,216.
Availability of data and materials
All data analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
UJ, EM, JV and IG participated in study design, interpreted the results and wrote the manuscript. UJ, MF, BB and FB involved in data acquisition and in laboratory work. YB, JK, TA, ND, AB, CH, LL participated in the review of literature. LA and AI provided critical revision of the manuscript. All authors approved final version of manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The author highlight that under Moroccan law no ethical approval is required for a retrospective study based on laboratory data and no consent from patients is necessary to carry out further tests in samples collected for other purposes. In our clinical bacteriology laboratory, the bacterial isolates routinely collected from various diagnostic and surveillance samples are stored at −20 °C. The present study thus is descriptive of a bacterial collection of those isolates as well as additional environmental isolates. All data were collected while maintaining anonymity. Ethical approval was therefore not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Samples sources, collection dates, wards, OXA-type carbapenemases and PFGE of A. baumannii isolates. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Uwingabiye, J., Lemnouer, A., Roca, I. et al. Clonal diversity and detection of carbapenem resistance encoding genes among multidrug-resistant Acinetobacter baumannii isolates recovered from patients and environment in two intensive care units in a Moroccan hospital. Antimicrob Resist Infect Control 6, 99 (2017). https://doi.org/10.1186/s13756-017-0262-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-017-0262-4