Abstract
Background
Acinetobacter baumannii is an important hospital-acquired pathogen in healthcare facilities that frequently causes bacteraemia and ventilator-associated pneumonia in intensive care units. Acinetobacter baumannii can be isolated from various sites in the hospital environment like medical equipment, bed linen, medical personnel and indwelling catheters. It is difficult to treat A. baumannii infections because of their highly resistant antimicrobial profiles. The purpose of this study was to determine the prevalence of β-lactamase genes in multidrug-resistant (MDR) clinical A. baumannii isolates using Multiplex-PCR (M-PCR) assays.
Methods
One hundred MDR A. baumannii isolates were collected from the diagnostic division of the Department of Medical Microbiology after routine analysis of the submitted specimens. All collected isolates were identified and tested for susceptibility using the VITEK 2® system (bioMérieux, France). Six isolates were excluded from this study because the isolates were incorrectly identified as A. baumannii with the VITEK 2® system (bioMérieux, France). Molecular tests, namely M-PCR assays, pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) were performed. MLST analyses were performed on representative isolates from the four major pulsotypes (≥5 isolates with 80 % similarity) and selective isolates from each minor pulsotype.
Results
All the A. baumannii isolates showed 100 % resistance to ampicillin, amoxicillin, cefuroxime, cefuroximine axetil, cefoxitin, cefotaxime and nitrofurantoin. Seven percent of the isolates were resistant to amikacin. Two percent of the isolates were classified as having intermediate susceptibility to tigecycline. A. baumannii isolates showed an antibiotic resistance profile of 67 % and higher to antibiotics, such as ceftazidime, cefepime, imipenem, meropenem, gentamicin, ciprofloxacin and trimethoprim/sulfamethoxazole. None of the isolates were resistant to colistin. The M-PCR assays showed that 99 % of the isolates contained the OXA-51 gene and 77 % contained the OXA-23 gene. None of the isolates contained the GES, GIM, IMP, KPC, NDM, OXA-24, OXA-58, PER, SIM, SPM, VEB and VIM genes. Representative A. baumannii isolates were grouped into five existing sequence types (ST): ST106, ST258, ST339, ST502, ST758 and ST848. Isolates belonging to the pan-European clonal lineages I and II (EUI and EUII) were identified.
Conclusion
The high prevalence of MDR A. baumannii isolates has a severe impact on available treatment choices and this in return impacts on treatment outcomes in the studied healthcare facilities. The most dominant ST among the collected isolates was ST758, member of the EUI group. The presence of the OXA-23 gene was not restricted to a specific ST. Continuous research and surveillance is necessary to monitor the circulating β-lactamase genes in clinical settings to guide infection control policies in order to try and curb the spread of this bacterium.
Similar content being viewed by others
Background
Acinetobacter baumannii is an emerging opportunistic pathogen that can become resistant to multiple antimicrobial agents [1–3]. Over the past 20 years A. baumannii’s clinical importance has been driven by its ability to obtain antimicrobial resistance factors [4, 5]. Resistance is acquired through the transfer of integrons, plasmids or transposons that carry groups of genes encoding resistance to several antibiotic families [6]. This bacterium can survive under a wide range of environmental conditions and on surfaces for extended periods of time and this can lead to both endemicity and outbreaks in healthcare facilities [4, 7].
Major problems are caused by this pathogen, especially among critically ill hospitalised patients in intensive care units (ICU’s) [5, 6, 8]. Clinical manifestations that can be associated with A. baumannii include: bacteraemia, meningitis, pneumonia, urinary tract infection, ventilator-associated pneumonia (VAP) and wound infections [8–10]. Patients with underlying chronic disease, decreased immunity, indwelling catheters and prolonged hospitalisation are more at risk to become colonised and develop subsequent infection with A. baumannii [7–9, 11].
This pathogen possesses different antimicrobial resistance mechanisms, which include the enzymatic inactivation of β-lactam antibiotics (cephalosporins, carbapenems and monobactams) through the production of extended spectrum β-lactamases, carbapenemases and AmpC-type (ampicillin class C) enzymes [12–15]. The β-lactamases of Gram-negative bacteria belong to Ambler classes A to D [12, 13]. The inherent class D oxacillinases (OXA) of A. baumannii belong to the chromosomally encoded OXA-51-like enzyme group [16–18]. However, the OXA-51 enzyme has been under considerable pressure due to antibiotic use and has evolved and now plays an important role in the carbapenem resistance of A. baumannii isolates [19]. If ISAbaI is located upstream of the bla OXA genes the A. baumannii isolate will have an increased OXA-51 expression which leads to carbapenem resistance (imipenem and meropenem) [19–21].
Infections caused by A. baumannii are difficult to treat because of the resistance to antimicrobial agents, especially β-lactams [3, 22]. There have been reports of MDR A. baumannii from hospitals all around the world (Argentina, Brazil, China, Europe and the United Kingdom) [4, 23, 24]. Only a few antimicrobial agents namely colistin (in combination with carbapenems) and tigecycline are available for the treatment of MDR A. baumannii [1, 2, 15]. However, there are now reports of colistin resistance in A. baumannii isolates, which is of great concern as colistin plays a pivotal role in MDR A. baumannii treatment [25–28]. Fortunately colistin resistance in A. baumannii is still rare [21, 25].
The aim of this study was to detect the prevalence of β-lactamase genes and to determine the genetic relationship among the A. baumannii isolates circulating in our clinical setting. The prevalence of β-lactamase genes is important for the surveillance of circulating A. baumannii strains as it provides more information for the infection control policies of the hospitals.
Methods
Study design and setting
The quantitative study was aimed to determine the prevalence of β-lactamase genes in consecutively collected A. baumannii isolates obtained from clinical specimens and to determine the genetic relatedness of circulating strains. The study was conducted at the Department of Medical Microbiology, Prinshof Campus, University of Pretoria/National Health Laboratory Service (NHLS) and received ethical approval from the Research Ethics Committee, Faculty of Health Sciences, University of Pretoria (121/2013); a waiver for informed consent was provided.
Collection, culturing and storage of A. baumannii clinical isolates
One hundred consecutive clinical A. baumannii isolates were collected without prior knowledge of patient information, hospital ward, specimen type and antibiotic susceptibility status. All 100 A. baumannii isolates were collected from the diagnostic laboratory during May to July 2013. The laboratory processes specimens from a tertiary academic hospital as well as district hospitals and various clinics as part of standard care. Routine analysis was performed as requested on the form and according to sample type. The isolates were identified and tested for susceptibility using the automated VITEK® 2 system (BioMérieux, France) with the VITEK® 2 GN card and the VITEK® 2 AST-N255 card. Gender, age, type of specimen, hospital, ward and the antibiotic susceptibility profiles were collected.
One colony of an isolate (obtained from a blood agar plate) was inoculated into 5 ml tryptone soya broth (TSB) (Oxoid, UK) and incubated on a shaking incubator (Vacutec, South Africa) at 35°C for a period of 18 to 24 h. Growth was indicated by turbidity in the broth. Isolates were stored by adding 900 μl of 50% glycerol (Merck, Germany) to 900 μl of the overnight TSB culture (Oxoid, UK) in 2 ml cryotubes (Greiner Bio-One, Germany) in a one to one ratio. This was done in triplicate and the isolates were stored in a -80°C freezer (New Brunswick, USA). The quality control strains used were the multidrug resistant A. baumannii ATCC® BAA-1605 strain; Klebsiella pneumoniae ATCC 8303 and K. pneumoniae ATCC BAA-2146.
DNA extraction of A. baumannii isolates
The genomic DNA of each of the 100 A. baumannii isolates was extracted from a single colony grown in TSB (Oxoid, UK) culture media by using the ZR Fungal/Bacterial kit [29]. The manufacturer’s instructions were followed with some modifications; instead of 50 to 100 mg of cells, 1 000 μl of overnight broth was used, centrifuged (Eppendorf 5417C, Germany) and the pellet was resuspended in phosphate buffered saline (PBS) solution (pH 7.2, Gibco® PBS, Life Technologies Corporation, USA) and 600 μl of the lysis solution were used instead of the prescribed 750 μl. The extracted genomic DNA was stored at -20 °C (Defy Ltd, Multimode, SA) until further analysis.
The detection of β-lactamase genes in A. baumannii by using M-PCR assays
Multiplex PCR assays were performed by using the QIAGEN® M-PCR kit (QIAGEN®, Germany) for the detection of several antibiotic resistance genes in A. baumannii (Table 1). An initial denaturation step of 95 °C for 15 min, followed by 30 cycles of denaturation at 94 °C for 30 s, an annealing temperature dependent on the melting temperature of the primer pair (multiplex I: 52 °C; multiplex II and III: 57 °C and multiplex IV: 60 °C) and extension at 72 °C for 90 s, followed by the final extension step at 70 °C for 10 min. A negative control (sterile ultrapure water) was included for all M-PCR assays. The following positive controls were used for multiplex I to IV: A. baumannii ATCC® BAA-1605™ strain; K. pneumoniae ATCC 8303 [Klebsiella pneumoniae carbapenemase (KPC) positive control] and K. pneumoniae ATCC BAA-2146 [New Delhi metallo-β-lactamase (NDM) positive control]. All isolates testing negative for the presence of the inherent OXA-51 gene were sent for identification using matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF).
Analysis of the Multiplex PCR products
A 1.8 % (m/v) MetaPhor agarose gel (Lonza, USA) with 5 μl of ethidium bromide (EtBr) (10 μg.ml−1) (Promega, USA) was used to visualise the PCR products after gel electrophoresis (Bio-Rad, USA). The 1.8 % (m/v) MetaPhor® agarose gel was prepared according to the manufacturer’s instructions (Lonza, USA).
A ready-to-use 100 bp DNA ladder (Fermentas, ThermoScientific, USA) was used to determine the approximate sizes of the amplicons. Gel electrophoresis (Bio-Rad, Germany) was performed at 90 V for 100 min with 1x Tris-Borate-EDTA (TBE) buffer [40 mM Tris-HCl, 20 mM Boric acid (Sigma-Aldrich, USA) and 1 mM EDTA (Sigma-Aldrich, USA), pH 8.0]. The bands were visualised and captured using a UVP Doc It transilluminator (Ultra-violet Products Incorporated, USA).
Determination of the genetic relationship of clinical A. baumannii isolates using the PFGE assay
The Pulsenet PFGE protocol [30] was followed with some modifications. The optical density (OD) was measured at 630 nm. The absorbance was adjusted to an OD range of 1.4 to 1.6 by using the ELx800 absorbance microplate reader (Bio-TEK, USA). The plugs were incubated overnight at 51°C and after incubation the plug was transferred to a new BD falcon tube (BD Biosciences, USA). The ApaI enzyme (New England Biolabs, USA) was used and the Salmonella choleraesuis serovar Braenderup (ATCC BAA-664) was used as a marker. Gel electrophoresis was performed using the following set conditions: interval 25 linear 5; angle 120° constant; voltage 220 V linear to 200 V and ran for 24 h. After the PFGE run was completed, the 1.2 % (m/v) SeaKem® LE agarose (Lonza, USA) gel was stained in 1 L distilled water, containing 250 μl EtBr (10 μg.ml-1) for 15 min and was destained in distilled water for 30 min. The gel was viewed and documented with a UV Doc It transilluminator (Ultra-violet Products Incorporated, USA). The banding patterns were analysed using the GelCompar II software programme (Applied Maths, Belgium). A distance matrix was constructed using the Dice coefficient and a dendogramme was constructed using the unweighted pair group method with arithmetic mean (UPGMA) [31]. Pulsotype designation was based on isolates showing ≥80 % relatedness, which corresponds to the Tenover criteria of possibly related 4 to 6 band differences between isolates [32, 33]. Representatives from each major PFGE pulsotype (≥5 isolates) and selective minor pulsotypes (< 5 isolates) with ≥80 % similarity were chosen for MLST analyses.
Molecular epidemiology
Seven housekeeping genes were used for MLST which included the following genes: (i) citrate synthase (gltA); (ii) DNA gyrase subunit B (gyrB); (iii) glucose dehydrogenase B (gdhB); (iv) homologous recombination factor (recA); (v) 60-kDa chaperonin (cpn60); (vi) glucose-6-phosphate isomerase (gpi) and (vii) RNA polymerase sigma factor (rpoD) (http://pubmlst.org/abaumannii/info/primers_Oxford.shtml). All the primers were synthesised by Inqaba Biotechnical Industries, South Africa.
All seven housekeeping genes were individually amplified in a 50 μl reaction as described in the Bioline kit’s guideline (Bioline, UK). The following programme was used on the thermal cycler (G-storm Model GS4822, Labtech, UK): an initial denaturation step of 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, an annealing temperature at 50 °C for 30 s, extension at 72 °C for 30 s, followed by the final extension step at 72 °C for 5 min [34]. The amplicons were visualised on a 1.8 % (m/v) MetaPhor agarose gel (Lonza, USA) as described previously. All the amplicons were sequenced in both forward and reverse directions by Inqaba Biotechnical Industries, South Africa. An ABI file obtained for each sequence were analysed with the CLC Main Workbench Version 6.0 (CLCbio, USA). The sequences were assigned to corresponding allelic profiles and sequence types by using the MLST Oxford database (http://pubmlst.org/abaumannii/).
Data analysis
One hundred consecutive A. baumannii isolates were collected, which were identified by the automated VITEK® 2 system (BioMérieux, France). Six isolates were excluded from this study because the isolates were incorrectly identified as A. baumannii by the VITEK® 2 system. These isolates were identified by MALDI-TOF analysis as: A. pittii; K. pneumoniae; Pseudomonas putida (2x); Staphylococcus aureus and S. epidermidis.
The results are reported on 94 clinical A. baumannii isolates. The proportions of A. baumannii isolates with Ambler classes A, B and D resistance genes were determined for the 94 isolates. The results were reported as proportions or percentages along with 95 % confidence intervals for the given sample size with an accuracy of within 10 %. The antimicrobial susceptibility and genotypic results obtained in this study were compared to a study done in 2008 in the same clinical setting [5]. The antimicrobial susceptibility profiles were also compared to the surveillance data from 12 sentinel public hospitals in South Africa (SA) [35].
Results
Collection of A. baumannii clinical isolates
Fifty-eight of the isolates were collected from male patients and 36 isolates were collected from female patients. Forty-three percent of the isolates were collected from diverse sites, 4 % from urine, 41 % from sputum and 12 % from blood cultures. The mean age of the patients was 39 years (ranging from 4 days to 77 years). A. baumannii was isolated from various wards but was the most prevalent in the medical and pulmonology ICU (12 %). Age and sex is not a cofounder and hence the odds ratio for area need not be adjusted. The crude odds ratio is 1.61 with a 95 % confidence interval (0.60; 4.31) and is not significant (p = 0.345).
Susceptibility profiles of A. baumannii isolates using the VITEK 2® system
All 94 of the A. baumannii isolates were resistant to ampicillin, amoxicillin, cefuroxime, cefuroximine axetil, cefoxitin, cefotaxime and nitrofurantoin. The A. baumannii isolates showed high level carbapenem and cephalosporin resistance (imipenem 86 %; meropenem 86 %; cefepime 90 % and ceftazidime 89 %). Table 2 shows the antimicrobial susceptibility profiles of the collected 94 A. baumannii isolates.
The 94 A. baumannii isolates showed an increase in antimicrobial resistance profiles compared to the study done by Kock et al. [5] in 2008: imipenem were 59 % in 2008 and 86 % in 2013 and meropenem were 63 % in 2008 and 86 % in 2013. The cephalosporin resistance patterns changed as follows: cefepime were 62 % in 2008 and were 90 % in 2013; ceftazidime were 45 % in 2008 and 89 % in 2013. Figure 1 shows the overall percentage of resistance in 2008 and 2013 in the Pretoria region. Data from the communicable diseases surveillance bulletin [35] showed that the A. baumannii isolates collected in 2013 in public hospitals in SA have resistance towards carbapenems and cephalosporins (imipenem 77 %; cefepime 80 % and ceftazidime 73 %).
Molecular identification of resistance genes found in A. baumannii isolates using M-PCR assays
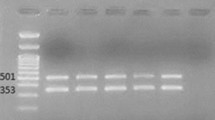
The M-PCR I assay showed that 99 % of the isolates contained the OXA-51 gene and 77 % contained the OXA-23 gene. None of the isolates were positive for OXA-24/40 and OXA-58. Figure 2 illustrates the M-PCR I assay, which targeted the OXA-23 (501 bp), OXA-24/40 (246 bp), OXA-51 (353 bp) and OXA-58 (599 bp) genes.
Results obtained after the M-PCR I assay, using a 1.8% (m/v) Metaphor agarose gel electrophoresis for the detection of oxacillinase (OXA) genes in A. baumannii isolates. The M-PCR assay amplified the OXA-23 (501 bp) and OXA-51 (353 bp) genes. Lanes 3, 5 and 7 were negative for all of the OXA genes. Lanes 2, 4, 8, 9, 10, 11 and 12 were positive for OXA-23 and OXA-51. Lane 6 was positive for OXA-51 only. Lane 1 was the negative control. Lane “L” represented the O’Range Ruler™ 100 bp DNA ladder
The results of the M-PCR assay I revealed the presence of two resistant profile types of A. baumannii that were circulating in the studied clinical setting. Type 1 was positive for both OXA-51 and OXA-23 (77 %) and type 2 was only positive for OXA-51 (22 %).
The M-PCR assay II [Imipenem metallo-β-lactamase (IMP), KPC and Verona integrin-encoded metallo-β-lactamase (VIM) genes]; III [Guyana extended-spectrum β-lactamase (GES), Pseudomonas extended resistance (PER) and Vietnam extended-spectrum β-lactamase (VEB) genes] and IV [German imipenemase (GIM), NDM-1, Seoul imipenem metallo-β-lactamase (SIM-1) and Sao Paulo metallo-β-lactamase (SPM) genes] were negative for all the tested genes.
Determination of the genetic relationship and global epidemiology of clinical A. baumannii isolates
The PFGE analysis showed that the 94 A. baumannii isolates can be grouped into four major pulsotypes (includes ≥ 5 isolates) and several minor pulsotypes (< 5 isolates) (Fig. 3). The representative A. baumannii isolates from each major pulsotype and selected minor pulsotypes were grouped into five existing STs (ST106, 1-1-1-1-1-98-6; ST258, 1-15-8-10-28-110-32; ST339, 44-73-4-11-44-121-4; ST502, 1-12-3-2-2-100-3; ST758, 1-17-8-10-28-106-32 and ST848, 1-15-3-2-2-142-3). Two groups (EUI: ST258, ST758; EUII: ST502, ST848) and singletons (ST106, ST339) which shared five out of seven alleles were identified with BURST analysis (http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_abaumannii_oxford_seqdef&page=listQuery&scheme_id=1&set_id=1).
PFGE patterns of A. baumannii isolates. The bandings patterns were analysed using the GelCompar II software programme (GelCompar II, Applied Maths, Belgium). A distance matrix was constructed using the Dice coefficient and a dendogramme was constructed from the distance matrix using UPGMA. Major pulsotype designation was based on five or more isolates showing ≥80 % relatedness. Isolate name, ST and the carriage of bla OXA-23 (Y, positive; N, negative) are indicated
Discussion
The increase in resistance profiles of A. baumannii can be due to the misuse of antibiotics in clinical settings, or because of patients who do not complete the prescribed antibiotic course. The bla OXA genes have an impact on treatment because it can already be resistant to the new inhibitors, such as β-lactamase inhibitors, or it can rapidly gain resistance [19].
The ability of the OXA β-lactamases to confer carbapenem resistance has already had a huge impact on the ability to treat Gram-negative pathogens and the current situation suggests that the problem is only set to increase [19]. The advantage of using molecular tests, such as the M-PCR assays used in this study, is that resistance genes circulating in clinical settings can be determined as this is not possible with phenotypic tests. Through molecular methods this study has indicated that 99 % of the A. baumannii isolates contained the OXA-51 gene, compared to Kock et al. [5] where only 81% out of the 97 isolates contained this gene. In the Kock et al. [5] study, the species identification obtained by the VITEK® 2 system was not confirmed by an alternative method, such as MALDI-TOF MS and thus possibly includes the other species of the A. baumannii-calcoaceticus complex. The results obtained in the current study shows the establishment of the species, A. baumannii in our clinical setting. The OXA-51 cluster is chromosomally encoded and is inherent to all A. baumannii isolates [4, 16, 19, 23, 35]. Seven isolates (of which six isolates were excluded) did not have the inherent OXA-51 gene. These isolates were further analysed using MALDI-TOF, which identified the seven isolates as: A. baumannii; A. pittii; K. pneumoniae; Pseudomonas putida (2x); Staphylococcus aureus and S. epidermidis. Studies have shown that the detection of the bla OXA-51-like gene can be used as a simple and reliable way of identifying A. baumannii [36, 37]. Although it is clear that bla OXA-51-like genes are present in at least the vast majority of isolates of A. baumannii, there has been some debate as to whether they are present in all isolates of this species [36]. Zander et al. [38] reported that three A. baumannii isolates did not harbour the predicted bla OXA-51-like gene (353 bp) and instead amplified PCR products of 1.2 kb and 1.6 kb. The authors then used external primers and the amplicons revealed that the one isolate (Ab-511) possessed the bla OXA-51 variant bla OXA-66, which was interrupted by the insertion sequence ISAba15 at nucleotide number 435, while the two other isolates (Ab-508 and Ab-653) possessed the bla OXA-51 variant bla OXA-78, which was disrupted at nucleotide number 379 by a novel insertion sequence ISAba19 [38]. Further investigation of the genomic environment of the isolate identified as A. baumannii by MALDI-TOF will reveal the changes in the OXA-51 genetic structure. Additionally the 16S-23S ribosomal spacer region can be sequenced for further species confirmation [37].
The occurrence of the OXA-23 gene has been reported since 1985 in A. baumannii isolates and has now disseminated worldwide [5, 19, 39, 40]. The prevalence of OXA-23 in this study was 77 %, compared to Kock et al. [5], who reported a 59 % prevalence in the same clinical setting in 2008. It is clear that the prevalence of OXA-23 in A. baumannii isolates in this clinical setting is increasing. This increased occurrence can be due to poor infection control practices, spread through contact with healthcare workers or from patient to patient and prolonged antibiotic use [11]. Stricter adherence to infection prevention and control policies by healthcare workers are needed to prevent further dissemination [4, 41, 42]. Correct use of disinfectants and solutions containing ethanol is particularly important because if applied at very low concentrations A. baumannii bacterium can become more pathogenic [41].
The high prevalence of OXA-23 can be due to the acquisition of genetic elements, such as plasmids and transposons [1, 42]. Liakopoulos et al. [43] conducted a study in 2010 to 2011 and reported a 95 % prevalence of OXA-23 in Greece. The OXA-23 cluster can be located on a plasmid or chromosome [5]. Mugnier et al. [44] reported in 2010 that eleven isolates had the bla OXA-23 gene on the chromosome, nine isolates had the bla OXA-23 gene on a plasmid and one isolate had two copies of the bla OXA-23 gene, one on the plasmid and one on the chromosome. The OXA-23 enzyme has a higher affinity to hydrolyse imipenem than meropenem, ertapenem or doripenem [19]. The MDR A. baumannii isolates in this study that contained the OXA-23 gene showed a 69 % resistance to imipenem (1 % had intermediate susceptibility and 6 % were susceptible to imipenem). Imipenem susceptibility may be explained by the absence of the ISAbaI element upstream of the bla OXA-23 gene [45]. Twenty-two percent of the isolates were positive for the OXA-51 gene only. However, 17 % of these isolates were resistant to imipenem while 5 % were susceptible.
The prevalence rates of isolates that contained both the OXA-23 and OXA-51 genes were 77 %. It is evident that the carbapenem-hydrolysing class D β-lactamases (CHDL’s), such as OXA-23 and OXA-51 were prevalent in clinical A. baumannii isolates in this study. The high prevalence of the OXA-51 and OXA-23 group (77 %) is due to the spread and establishment of several successful clones in our clinical setting.
None of the clinical A. baumannii isolates contained the genes that were screened for in M-PCR II to IV as well as the OXA-24 and OXA-58 genes that were part of M-PCR I. The GIM, SIM and SPM metallo-β-lactamase (MBL) genes are rarely found in A. baumannii isolates [46]. Kock et al. [5] reported one A. baumannii isolate that was positive for the VIM-like gene. Safari et al. [47] reported that 99 % of the isolates in ICU wards of three educational hospitals of Hamadan City in Iran produced metallo-β-lactamases. In 2013, the first outbreak of NDM-1 producing A. baumannii was reported in France, which occurred in a surgical ICU [48]. Fortunately NDM-1 in A. baumannii has not yet been detected in our clinical setting. The NDM-1 gene encodes an MBL carbapenemase that has a high hydrolytic activity for carbapenem antibiotics, and much more [49].
PFGE is suited for studying isolates from a defined temporal and spatial epidemiologic setting, whereas MLST has an advantage in defining clonal lineages of isolates from larger geographic areas over time [50]. Three clonal lineages of A. baumannii, commonly referred to as the pan-European clonal lineages (EUI, EUII, and EUIII), have predominated in many European countries since the 1990s [50]. Isolates belonging to the pan-European clonal lineages I and II (EUI and EUII) were identified in our clinical setting. EUI (ST258 and ST758) was predominant together with the OXA-23 gene. Other diverse clones, such as ST106 and ST339 are also circulating in our clinical setting. The origin of the A. baumannii isolates in our setting cannot be attributed to the dissemination of one or two successful clones but rather to the dissemination of a diverse group of successful clones.
Conclusions
The increasing resistance to carbapenems are of great concern due to the limited number of effective drugs and the association with increased morbidity and mortality. There was a noticeable change in the resistance profiles of the A. baumannii isolates from 2008 to 2013 in the same setting. The high prevalence of MDR A. baumannii isolates has a severe impact on treatment outcomes in the studied healthcare facilities. The presence of the OXA-23 gene was not related to a specific ST and can be due to the acquisition of genetic elements, such as plasmids and transposons. OXA-23 and ST758 were dominant among the isolates. A limitation of this study is the small sample size and only tertiary academic hospitals in the Pretoria region were included and thus the findings in this study are not a true reflection of the situation in the whole of SA. Another limitation of this study is that there were not any positive controls included for the screened genes in M-PCR assay III, which could have caused that these genes were missed due to not using the optimal PCR conditions.
It is vital to identify and characterise healthcare-associated pathogens and their circulating antibiotic resistance genes as it is of great concern to public health systems. Understanding the resistance genes can lead to the understanding of their origin and can assist in better infection control measures and ultimately limit the spread of these pathogens.
Abbreviations
- AmpC:
-
Ampicillin class C
- CHDL:
-
Carbapenem-hydrolysing class D β-lactamase
- EtBr:
-
Ethidium bromide
- GES:
-
Guyana extended-spectrum β-lactamase
- GIM:
-
German imipenemase
- ICU:
-
Intensive care unit
- IMP:
-
Imipenem metallo-β-lactamase
- KPC:
-
Klebsiella pneumoniae carbapenemase
- MALDI-TOF:
-
Matrix assisted laser desorption ionization-time of flight mass spectrometry
- MBL:
-
Metallo-β-lactamase
- MDR:
-
Multidrug resistant
- MLST:
-
Multilocus sequence typing
- M-PCR:
-
Multiplex polymerase chain reaction
- NDM:
-
New Delhi metallo-β-lactamase
- NHLS:
-
National health laboratory services
- NRF:
-
National Research Foundation
- OD:
-
Optical density
- OXA:
-
Oxacillinases
- PBS:
-
Phosphate buffered saline
- PER:
-
Pseudomonas extended resistance
- PFGE:
-
Pulsed field gel electrophoresis
- SA:
-
South Africa
- SIM:
-
Seoul imipenem metallo-β-lactamase
- SPM:
-
Sao Paulo metallo-β-lactamase
- ST:
-
Sequence type
- TBE:
-
Tris-Borate-EDTA
- TSB:
-
Tryptone soya broth
- VAP:
-
Ventilator-associated pneumonia
- VEB:
-
Vietnam extended-spectrum β-lactamase
- VIM:
-
Verona integrin-encoded metallo-β-lactamase
References
Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, Muto CA, et al. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob Agents Chemother. 2008;52:3837–43.
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 2010;5:1–17.
Gbaguidi-Haore H, Dumartin C, L’Hériteau F, Péfau M, Hocquet D, Rogues A, et al. Antibiotics involved in the occurrence of antibiotic-resistant bacteria: a nationwide multilevel study suggests differences within antibiotic classes. J Antimicrob Chemother. 2013;68:461–70.
Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82.
Kock MM, Bellomo AN, Storm N, Ehlers MM. Prevalence of carbapenem resistance genes in Acinetobacter baumannii isolated from clinical specimens obtained from an academic hospital in South Africa. South Afr J Epidemiol Infect. 2013;28:28–32.
Fournier P, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:0062–72.
Maragakis LL, Perl TM. Acinetobacter baumannii: Epidemiology, antimicrobial resistance and treatment options. Clin Infect Dis. 2008;46:1254–63.
Chuang Y, Cheng C, Sheng W, Sun H, Wang J, Chen Y, et al. Effectiveness of tigecycline-based versus colistin-based therapy for treatment of pneumonia caused by multidrug-resistant Acinetobacter baumannii in a critical setting: a matched cohort analysis. BMC Infect Dis. 2014;14:102–9.
Javed A, Zafar A, Ejaz H, Zubair M. Frequency and antimicrobial susceptibility of Acinetobacter species isolated from blood samples of paediatric patients. Pak J Med Sci. 2012;28:363–6.
Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:233–8.
Cisneros JM, Rodríquez-Baño J. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin Microbiol and Infect. 2002;8:687–93.
Livermore DM, Woodford N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. TRENDS Microbiol. 2006;14:413–20.
Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–58.
Maamoun HAH. Molecular characteristics of extended-spectrum beta-lactamases among gram-negative isolates collected in Cairo University hospital. Comp Clin Pathol. 2013;22:733–9.
Sonnevend A, Ghazawi A, Al Munthari N, Pitout M, Hamadeh MB, Hashmey R, et al. Characteristics of epidemic and sporadic strains of Acinetobacter baumannii isolated in Abu Dhabi hospitals. J Med Microbiol. 2013;62:582–90.
Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents. 2010;35:219–26.
Chen T, Lee Y, Kuo S, Hsueh P, Chang F, Siu L, et al. Emergence and distribution of plasmids bearing the bla OXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother. 2010;54:4575–81.
Héritier C, Poirel L, Fournier P, Claverie J, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobater baumannii. Antimicrob Agents Chemother. 2005;49:4174–9.
Evans BA, Amyes SGB. OXA β-lactamases. Clin Microbiol Rev. 2014;27:241–63.
Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, et al. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:1589–93.
Agodi A, Voulgari E, Barchitta M, Quattrocchi A, Bellocchi P, Poulou A, et al. Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two sicilian hospitals. J Hosp Infect. 2014;208:1142–51.
Constantiniu S, Romaniuc A, Iancu LS, Filimon R, Taraşi I. Cultural and biochemical characteristics of Acinetobacter spp strains isolated from hospital units. Am J Prev Med. 2004;12:35–42.
Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84.
Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63:55–9.
Lee SY, Shin JH, Park KH, Kim JH, Shin MG, Sug SP, et al. Identification, genotypic relation, and clinical features of colistin-resistant isolates of Acinetobacter genomic species 13BJ/14TU from bloodstreams of patients in a university hospital. J Clin Microbiol. 2014;52:931–9.
Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, et al. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58:828–32.
Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the PmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55:3370–9.
Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, et al. Emergence of colistin-resistant in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J Infect Dis. 2013;208:1142–51.
Thermo Scientific, 2013: Simple, rapid isolation of DNA from tough-to-lyse fungi and bacteria. www.zymoresearch.com/downloads/dl/file/id/215/d6007d.pdf. 2013
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67.
Fillaux J, Dubouix A, Concil J, Laguerre J, Marty N. Retrospective analysis of multidrug-resistant Acinetobacter baumannii strains isolated during a 4-year period in a university hospital. Infect Control Hosp Epidemiol. 2006;27:647–53.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by Pulsed-Field Gel Electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, et al. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother. 2014;58:3762–7.
Institut Pasteur: Acinetobacter baumannii MLST Database. Available: http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html Last full update: 23 October 2014 Accessed: January 2015
Perovic O, Fortuin-de Smidt M, Chetty V. Antimicrobial resistance surveillance from sentinel public hospitals, South Africa, 2013. Comm Dis Surveil Bull. 2014;12:30–58.
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the bla OXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–6.
Merkier AK, Centrón. bla OXA-51-type β-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int J Antimicrob Agents. 2006;28:110–3.
Zander E, Higgins PG, Fernández-González, Seifert H. Detection of intrinsic bla OXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int J Med Microbiol. 2013;303:88–9.
Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–9.
Coelho J, Woodford N, Afzal-Shah M, Livermore D. Occurrence of OXA-58-like carbapenemases in Acinetobacter spp. Collected over 10 years in three continents. Antimicrob Agents Chemother. 2006;50:756–8.
Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–62.
Fournier P, Richet H. The Epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692–9.
Liakopoulos A, Miriagou V, Katsifas EA, Karagouni AD, Daikos GL, Tzouvelekis LS, et al. Identification of OXA-23-producing Acinetobacter baumannii in Greece, 2010 to 2011. Euro Surveil. 2012;17:1–3.
Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the bla OXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40.
Carvalho KR, Carvalho-Assef APD, dos Santos LG, Pereira MJF, Asensi MD. Occurrence of bla OXA-23 gene in imipenem-susceptible Acinetobacter baumannii. Mem Inst Oswaldo Cruz. 2011;106:505–6.
Nordmann P, Poirel L. Acinetobacter baumannii: basic and emerging mechanisms of resistance. Eur Infect Dis. 2008;2:94–7.
Safari M, Saidijam M, Bahador A, Jafari R, Alikhani MY. High prevalence of multidrug resistance and metallo-β-lactamase (MβL) producing Acinetobacter baumannii isolated from patients in ICU wards, Hamadan, Iran. J Res Health Sci. 2013;13:162–7.
Decousser JW, Jansen C, Nordmann P, Emirian A, Bonnin RA, Anais L, et al. Outbreak of NDM-1-producing Acinetobacter baumannii in France, January to May 2013. Euro Surveill. 2013;18:1–4.
Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y. Epidemiological characteristics and genetic structure of bla NDM-1 in non-baumannii Acinetobacter spp. in China. J Antimicrob Chemother. 2012;67:2114–22.
Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian G, Marschall J, Urban CM, et al. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol. 2011;49:3849–54.
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–3.
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–5.
Voets GM, Fluit AC, Scharringa J, Stuart JC, Leverstein-van Hall MA. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int J Antimicrob Agents. 2011;37:356–9.
Acknowledgements
The authors would like to thank the Department of Medical Microbiology and the NHLS for the use of their facilities and for the provided isolates. The authors also want to thank Dr Anthony Smith, coordinator for PulseNet Africa for the donation of the Salmonella strain (ATCC BAA-664). ML was supported by a National Research Foundation (NRF) grant. The MALDI-TOF analysis is based on research supported in part by the National Research Foundation (NRF) of South Africa (Grant specific unique reference number (UID) 74426). Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF. The statistical analysis was performed with the assistance of Prof PJ Becker.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ML, MME, AWD and MMK drafted and finalised the manuscript. ML collected all the clinical A. baumannii isolates and carried out the laboratory assays. ML, MME, AWD and MMK participated in data analysis and interpretation. ML, MME and MMK were involved in the concept design. All authors read and approved the final version of the manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lowings, M., Ehlers, M.M., Dreyer, A.W. et al. High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa – an update. BMC Infect Dis 15, 521 (2015). https://doi.org/10.1186/s12879-015-1246-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-015-1246-8