Abstract
Background
Apicomplexan tick-borne pathogens that cause disease in companion animals include species of Babesia Starcovici, 1893, Cytauxzoon Neitz & Thomas, 1948, Hepatozoon Miller, 1908 and Theileria Bettencourt, Franca & Borges, 1907. The only apicomplexan tick-borne disease of companion animals that is known to occur in Australia is babesiosis, caused by Babesia canis vogeli Reichenow, 1937 and Babesia gibsoni Patton, 1910. However, no molecular investigations have widely investigated members of Apicomplexa Levine, 1980 in Australian ticks that parasitise dogs, cats or horses, until this present investigation.
Results
Ticks (n = 711) removed from dogs (n = 498), cats (n = 139) and horses (n = 74) throughout Australia were screened for piroplasms and Hepatozoon spp. using conventional PCR and Sanger sequencing. The tick-borne pathogen B. vogeli was identified in two Rhipicephalus sanguineus Latreille ticks from dogs residing in the Northern Territory and Queensland (QLD). Theileria orientalis Yakimov & Sudachenkov, 1931 genotype Ikeda was detected in three Haemaphysalis longicornis Neumann ticks from dogs in New South Wales. Unexpectedly, the exotic tick-borne pathogen Hepatozoon canis James, 1905 was identified in an Ixodes holocyclus Neumann tick from a dog in QLD. Eight novel piroplasm and Hepatozoon species were identified and described in native ticks and named as follows: Babesia lohae n. sp., Babesia mackerrasorum n. sp., Hepatozoon banethi n. sp., Hepatozoon ewingi n. sp., Theileria apogeana n. sp., Theileria palmeri n. sp., Theileria paparinii n. sp. and Theileria worthingtonorum n. sp. Additionally, a novel cf. Sarcocystidae sp. sequence was obtained from Ixodes tasmani Neumann but could not be confidently identified at the genus level.
Conclusions
Novel species of parasites in ticks represent an unknown threat to the health of companion animals that are bitten by these native tick species. The vector potential of Australian ticks for the newly discovered apicomplexans needs to be assessed, and further clinical and molecular investigations of these parasites, particularly in blood samples from dogs, cats and horses, is required to determine their potential for pathogenicity.
Similar content being viewed by others
Background
Intracellular apicomplexan parasites consist of the groups haemococcidia, haemogregarines, haemosporidia and piroplasms; collectively these haemoprotozoa are transmitted by haematophagous vectors, such as ticks (Acari: Ixodida). Apicomplexan tick-borne diseases of companion animals are caused by the piroplasms Babesia spp., Cytauxzoon felis Kier, 1979 and Theileria spp., and the haemogregarines Hepatozoon Miller, 1908 spp. Unlike other developed countries with high pet ownership rates, such as the USA [1], relatively few apicomplexan tick-borne pathogens have been reported to affect pets in Australia; these include Babesia canis vogeli Reichenow, 1937 [2, 3] and Babesia gibsoni Patton, 1910 [4], which cause babesiosis in domestic dogs (Canis lupus familiaris Linnaeus). The brown dog tick (Rhipicephalus sanguineus Latreille), a species that was introduced into the Australian continent in relatively recent times, is the vector of B. vogeli [5, 6] and a putative vector of B. gibsoni [4]. Although the majority of ticks collected from dogs during a 2012–2015 national tick survey in Australia were identified as R. sanguineus (73%) [7], seven out of the ten tick species identified on dogs were native ticks that, to the best of our knowledge, have not been widely investigated for their association with apicomplexan parasites previously. Similarly, most of the tick species that were identified on horses (Equus ferus caballus Linnaeus), and all of those removed from cats (Felis catus Linnaeus), were also native ticks [7]. To gain a more comprehensive understanding of the potentially vector-borne parasites associated with Australian ticks, the current study aimed to screen ticks that were parasitising dogs, cats and horses for piroplasms and Hepatozoon spp. using conventional PCR and Sanger sequencing.
Methods
Tick collection and identification
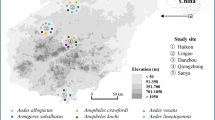
Ticks were opportunistically collected during a nationwide tick survey between 2012–2015 (n = 4765) [7]. Individual specimens were removed from companion animals, stored in 70% ethanol and morphologically identified [8, 9]. A subset of ticks (n = 711) was selected for the present study from dogs (n = 498), cats (n = 139) and horses (n = 74) (Table 1). Collection locations included all Australian states and territories, except for the Australian Capital Territory. A summary of the tick collection locations is provided in Additional file 1: Table S1.
DNA extractions
Tick specimens were bisected and genomic DNA (gDNA) was extracted from one half of the tick (cut into smaller pieces with sterile scalpel blades; the other half of the specimen was stored in ethanol for future studies) using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations, with the following modifications: after the addition of buffer ATL and proteinase K, the 56 °C incubation time was increased to ~16 h, i.e. samples were incubated overnight; the volume of elution buffer AE was decreased to 50 μl to increase the gDNA concentration; and the elution step was repeated on the 50 μl eluate to increase the gDNA yield. Extraction reagent blank controls (ExCs) (n = 21) were included alongside each batch of gDNA extractions.
PCR amplification
Initial piroplasm and Hepatozoon spp. screening
The 711 gDNA samples from ticks and 21 ExCs were initially screened for a short 300 bp region of the 18S ribosomal RNA gene (18S) of piroplasms and Hepatozoon spp. with the primers 18SApiF/18SApiR (Table 2) that were designed in Geneious v10.2.2 [10] based on an alignment of piroplasms and Hepatozoon species. Conventional PCRs were performed in 25 μl reaction volumes with 1× KAPA Taq buffer (Sigma-Aldrich, St. Louis, Missouri, USA), 2 mM MgCl2, 1 mM dNTPs, 0.04 mg BSA (Fisher Biotec, Perth, Western Australia, Australia), 400 nM of each forward and reverse primer, 0.02 U KAPA Taq DNA Polymerase (Sigma-Aldrich) and 1 μl of neat gDNA. Thermal cycling conditions included an initial denaturation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 45 s, with a final extension of 72 °C for 5 min. No-template controls (NTCs) were included alongside all PCR assays.
Babesia spp. and Theileria spp. amplification
All samples that were 18SApiF/18SApiR-positive were sequenced using Sanger sequencing and apicomplexan species were identified using the Basic Local Alignment Search Tool (BLAST) for sequence comparison to the National Centre for Biotechnology Information (NCBI) non-redundant nucleotide (nr/nt) database (methods described in later sections). Samples that were positive for B. vogeli, Hepatozoon spp. or had mixed sequence chromatograms from the 18SApiF/18SApiR PCR screening were then subjected to a nested Babesia spp. and Theileria spp. PCR assay using the external primers BTF1/BTR1 that target a 930 bp region of 18S and the internal primers BTF2/BTR2 that target an 800 bp region of 18S [11] (Table 2). The PCR assays were carried out according to the 18SApiF/18SApiR PCR assay conditions described in the present study, with the following modification: the final MgCl2 concentration was 1.5 mM. The thermal cycling conditions for the BTF1/BTR1 and BTF2/BTR2 primer sets were carried according to previously described methodologies [11], with the following modification: the denaturation temperature was increased to 95 °C for BTF1/BTR1 and BTF2/BTR2.
Following unsuccessful attempts to achieve amplification of a long (> 1300 bp) region of 18S for all samples that were positive for novel Babesia and Theileria species with the previously published primer sets BT18SF1/BT18SR1 and BT18SF2/BT18SR2 (nested PCR) [12], and Nbab_1F [13] and TB-Rev [14], different primer combinations were tested that had similar melting temperatures (≤ 5 °C). The primer combinations included BTF1/TB-Rev [11, 15], BTF1/BT18SR2 [11, 12] and Nbab_1F/18SApiR [12, 13]. PCR assays were carried out according to the methods described for 18SApiF/18SApiR, with the following modifications: reaction volumes were increased to 50 μl and 2 μl of Theileria orientalis Yakimov and Sudachenkov, 1931 genotype Ikeda positive control gDNA was used. Thermal cycling conditions included an initial denaturation at 95 °C for 5 min followed by 50 cycles of denaturation at 95 °C for 30 s, annealing temperature (Tann) gradients were carried out ranging from 48–60 °C for 30 s and extension at 72 °C for 2 min, with a final extension of 72 °C for 5 min. Amplification of a ~1500 bp product was observed in the T. orientalis genotype Ikeda positive control for the Nbab_1F/18SApiR primers with a Tann of 60 °C (Table 2). These methods, with the Nbab_1F/18SApiR primers and Tann of 60 °C, were then used to amplify samples positive for novel Babesia and Theileria species using conventional PCR.
Hepatozoon spp. amplification
Samples that were positive for piroplasms, Hepatozoon canis James, 1905 and those that had mixed sequence chromatograms from the 18SApiF/18SApiR screening were then subjected to Hepatozoon spp. PCR assays using two different primer sets; HepF300/Hep900 [16] and HEMO1/HEMO2 [17] that target a 600 bp and 900 bp region of 18S, respectively (Table 2). The PCR assays for the HepF300/Hep900 primers were carried out according to the methods described for the 18SApiF/18SApiR primers in the present study, with the following modification: the final MgCl2 concentration was decreased to 1.5 mM. The thermal cycling conditions were followed according to previously published methods [16], with the following modifications: the denaturation temperature was increased to 95 °C; the number of cycles was increased to 40; and the final extension time was decreased to 5 min. For the HEMO1/HEMO2 primer set, the PCR assays and thermal cycling conditions were carried out using the 18SApiF/18SApiR methods described in the present study, with the following modifications: the number of cycles was increased to 45; the optimal Tann was determined by a Tann gradient to be 48 °C; and a 1 min extension time was used during the 45 cycles.
A ~1650 bp 18S region of the novel Hepatozoon species was amplified using the HAM-1F/HPF-2R primer set [18] (Table 2). The PCR assay and thermal cycling conditions described above for the Nbab_1F/18SApiR primers were used, but with a Tann of 56 °C.
Theileria orientalis genotyping
To determine the genotypes of T. orientalis, the Ts-U/Ts-R primers [19] were used to amplify 800 bp of the major piroplasm surface protein (MPSP) gene of T. orientalis that encodes MPSP p32 (Table 2). The PCR assays were carried out according to the 18SApiF/18SApiR methods described in this study. The thermal cycling conditions were followed according to previously published methods [15], with the following modifications: the denaturation temperature was increased to 95 °C; the number of cycles was increased to 40; and the final extension time was increased to 5 min.
Gel electrophoresis and PCR product purification
The amplified DNA was electrophoresed in 1% agarose gel containing SYBR Safe Gel Stain (Invitrogen, Carlsbad, California, USA) and visualized with a dark reader trans-illuminator (Clare Chemical Research, Dolores, Colorado, USA). PCR products of the expected amplicon size were excised from the gel with sterile scalpel blades and purified for Sanger sequencing using a filtered pipette tip method [20].
Sanger sequencing
Purified PCR products were sequenced in forward and reverse directions independently on a 96-capillary 3730xl DNA Analyzer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) using an ABI Prism™ BigDye v3.1. Cycle Sequencing kit (Applied Biosystems, Foster City, California, USA) according to the manufacturer’s instructions.
Phylogenetic analyses
Forward and reverse sequence chromatograms were aligned and merged to generate consensus sequences and were trimmed of primers using Geneious v10.2.2. BLAST was used to compare the consensus sequences to the NCBI nr/nt database. For phylogenetic analyses of piroplasm, Hepatozoon and the Coccidiasina: Eucoccidiorida, cf. Sarcocystidae sp. consensus sequences, the longest available 18S sequences on GenBank for named Piroplasmida, Adeleorina and Sarcocystidae species were imported into Geneious v10.2.2 and aligned using the MUSCLE alignment tool [21]. As partial 18S sequence lengths varied, the alignments were trimmed to retain as many named species as possible in overlapping hypervariable regions, but some sequences were removed from the alignment due to either their short length, or the region did not overlap with the majority of other sequences.
Phylogenetic analyses of piroplasm and Hepatozoon consensus sequences were carried out that also included GenBank sequences with ≥ 95% and ≥ 96% similarity, respectively, over a greater nucleotide alignment length. After sequences in these alignments were trimmed to the length of the shortest sequence with ≥ 95% or ≥ 96% similarity, duplicate sequences were removed. An alignment of Eucoccidiorida 18S sequences was generated to phylogenetically assess the grouping of the consensus cf. Sarcocystidae sp. sequence respective to other families of the Eucoccidiorida, and this alignment was trimmed to the length of the consensus cf. Sarcocystidae sp. sequence (572 bp).
Nucleotide alignments were imported into the program PhyML [22] and assessed for the most appropriate nucleotide substitution model based on Bayesian Information Criterion (BIC) and Bayesian phylogenetic trees were constructed using MrBayes v3.2.6 [23].
Sequences generated from this study have been submitted to GenBank under the accession numbers MG062865, MG571580-MG571582, MG593271-MG593276, MG758109-MG758121 and MG758124-MG758138.
Genetic distance estimates
The longer 18S fragment sequences from the novel species found in this study were compared to the NCBI nr/nt database using BLAST, and the 18S sequences from the most closely related unnamed and named species were imported into Geneious v10.2.2 for genetic pairwise distance (percent sequence identity) comparisons. The 18S sequences of the most closely related named species were then compared to the NCBI nr/nt database using BLAST, and the 18S sequences with the highest percent similarity to the named species were also imported into Geneious v10.2.2 for pairwise distance comparisons. As partial 18S sequences were obtained in the present study, the effect of shorter versus longer 18S alignments on pairwise distance estimates was assessed by comparing the percentage differences in pairwise identities between shorter (~1500 bp) and longer (~1650 bp) alignments of the most closely related named species. The program MUSCLE was used to build the alignments and the pairwise percent identities were calculated with a Kimura distance matrix [21].
Results
Prevalence of Apicomplexa species
Approximately 300 bp of 18S was amplified in 41/711 samples using the 18SApiF/18SApiR primer set; however, only 16/41 of these 18SApiF/18SApiR-positive samples had good quality chromatograms, while the rest had mixed chromatograms as a result of amplification of multiple eukaryotic organisms. No amplification was observed in the ExCs or NTCs for any of the PCR assays. The sequence accession numbers, lengths and top BLAST matches for all sequences obtained in this study are summarised in Additional file 2: Table S2.
Of the 26 samples for which mixed chromatograms were obtained using the 18SApiF/18SApiR primer set, Hepatozoon spp. PCRs yielded no amplification. Amplification with the BTF1/BTR1 and BTF2/BTR2 primer set identified one positive from the 26 samples (Haemaphysalis longicornis Neumann nymph, sample HLN3) that had mixed chromatograms. Additionally, mixed infections with Hepatozoon and Theileria species were detected in two Ixodes tasmani Neumann samples (Hepatozoon banethi n. sp. and Theileria apogeana n. sp. in sample ITF7; H. banethi n. sp. and Theileria palmeri n. sp. in sample ITF6) at an overall prevalence of 0.3% (2/711; 95% CI: 0–1.0%). A mixed infection with Theileria species (T. palmeri n. sp. and Theileria paparinii n. sp.) was detected in one I. tasmani sample (ITF1) (0.1%; 1/711; 95% CI: 0–0.8%). The overall prevalence of the Apicomplexa species in different states and territories, and Australia-wide, in all tick species from all host species is summarised in Table 3. The prevalence of Apicomplexa species Australia-wide and in each state and territory is summarised for all tick species from all hosts, individual tick species from all hosts, and individual tick species from dogs, cats and horses in Additional file 3: Table S3.
Overall, of the 41 samples that were positive by PCR, sequences were unambiguously confirmed in 17/711 samples (2.4%; 95% CI: 1.4–3.8%). The overall prevalence of the 12 Apicomplexa species (including the three mixed infections) based on confirmed sequences was 2.8% (20/711; 95% CI: 1.4–3.8%) (Table 3).
Endemic tick-borne pathogens: B. vogeli and T. orientalis
Two known endemic tick-borne pathogens were identified: B. vogeli and T. orientalis genotype Ikeda. Babesia vogeli (~300 bp) (100% homology) was detected in 1.1% of R. sanguineus ticks collected from dogs (2/184; 95% CI , 0.1–3.9%) (Additional file 3: Table S3); an R. sanguineus female from a dog in Queensland (QLD) (sample RSF1; MG758129) (3%; 1/33); 95% CI: 0.1–15.8%), and an R. sanguineus larva from a dog in the Northern Territory (NT) (sample RSL1; MG758131) (2%; 1/50; 95% CI: 0.1–10.6%) (Additional file 3: Table S3). Further characterisation of longer 18S sequences obtained from RSF1 (MG758130) and RSL1 (MG758132) with the nested piroplasm PCR assay confirmed their 100% homology to B. vogeli isolates in GenBank (Additional file 2: Table S2).
Theileria orientalis genotype Ikeda (100% homology) was detected in three H. longicornis nymphs at the 18S locus (HLN1-3) (MG571580-MG571582) at a prevalence of 4% (3/75; 95% CI: 0.8–11.2%) (Additional file 3: Table S3). These Theileria orientalis genotype Ikeda-positive H. longicornis nymphs were removed from dogs in New South Wales (NSW) (6.4%; 3/47; 95% CI: 1.3–17.5%) (Additional file 3: Table S3). Amplification of these samples at the MPSP p32 gene locus produced 835 bp sequences (MG758109-MG758111), which were 100% identical to each other, and 100% identical to Theileria orientalis strain Shintoku, genotype Ikeda (XM_009691550) (Additional file 2: Table S2).
An exotic tick-borne pathogen: H. canis
Unexpectedly, a 303 bp sequence with 100% sequence similarity to the exotic tick-borne pathogen, H. canis, was obtained from one engorged Ixodes holocyclus Neumann female (sample IHF2; MG062865) (0.3%; 1/335; 95% CI: 0–1.7%) removed from a dog in QLD (1.7%; 1/60; 95% CI: 0–8.9%). Further characterisation of this sample using the HepF300/Hep900 and HEMO1/HEMO2 primer sets produced sequences with a ~30 bp overlap, and this concatenated sequence (MG758124) was 1409 bp in length and was 99.9% similar, with two single nucleotide polymorphisms (SNPs), to H. canis (KX712124) (Additional file 2: Table S2).
Novel Babesia species
A novel Babesia species, Babesia mackerrasorum n. sp., was identified in a cf. Haemaphysalis Koch sp. male from a horse in NSW (sample HspM1; MG593271 and MG593276) (33.3%; 1/3; 95% CI: 0.8–90.6%) with 98.3% similarity to Babesia macropus Dawood, 2013 (JQ437265) isolated from an eastern grey kangaroo (Macropus giganteus Shaw) in NSW, Australia. Another novel Babesia species, Babesia lohae n. sp., was identified in an I. holocyclus female from a cat in QLD (sample IHF1; MG593272 and MG593273) (2%; 1/51; 95% CI: 0–10.4%), which was only 96.7% similar to B. mackerrasorum n. sp. (MG593271) (refer to pairwise genetic distance matrix in Additional file 4: Table S4), and 100% similar to Babesia sp. (MG251436) isolated from I. tasmani collected from a brushtail possum (Trichosurus vulpecula Kerr) in Australia (Additional file 2: Table S2).
Novel Hepatozoon species
Three I. tasmani ticks removed from dogs in TAS contained a novel Hepatozoon species; H. banethi n. sp. [samples ITF2 (MG758133 and MG758134), ITF6 (MG758135 and MG758136) and ITF7 (MG758138 and MG758137)] (6.8%; 3/44; 95% CI: 1.4–18.7%) (Additional file 3: Table S3). The long 18S H. banethi n. sp. sequences from ITF6 (MG758136) and ITF7 (MG758137) were 99.9% similar to each other, and the long 18S H. banethi n. sp. sequence from ITF2 (MG758133) was 99.8% similar to the sequences from ITF6 and ITF7 (Additional file 4: Table S4). The top NCBI BLAST results revealed that the long 18S sequences of H. banethi n. sp. were most similar (98.0% for ITF2 and ITF6, and 97.8% for ITF7) to Hepatozoon sp. (FJ719813) isolated from a colocolo opossum (Dromiciops gliroides Thomas) in Chile (Additional file 2: Table S2). Hepatozoon ewingi n. sp. was detected in Haemaphysalis bancrofti Nuttall and Warburton from a horse in NSW (sample HBM1; MG593274 and MG593275) (33.3%; 1/3; 95% CI: 0.8–90.6%) (Additional file 3: Table S3). The long 18S sequence of H. ewingi n. sp. (MG593275) was only 94.8% similar to H. banethi n. sp. sequences from ITF2 (MG758133) and ITF6 (MG758136) and 94.5% similar to the H. banethi n. sp. sequence from ITF7 (MG758137) (Additional file 4: Table S4), and was also most similar (96.3%) to Hepatozoon sp. (FJ719813) isolated from D. gliroides (Additional file 2: Table S2).
Novel Theileria species
Four novel Theileria species were found in I. tasmani ticks from dogs in Tasmania (TAS): T. apogeana n. sp. from sample ITF7 (MG758116 and MG758126) (2.3; 1/44; 95% CI: 0.1–12.0%); T. palmeri n. sp. from samples ITF1 (MG758113) and ITF6 (MG758120 and MG758125) (4.5%; 2/44; 95% CI: 0.6–15.5%); T. paparinii n. sp. from samples ITF1 (MG758112) and ITF4 (MG758115 and MG758117) (4.5%; 2/44; 95% CI, 0.6–15.5%); and Theileria worthingtonorum n. sp. from samples ITF3 (MG758114 and MG758118) and ITF5 (MG758119 and MG758121) (4.5%; 2/44; 95% CI: 0.6–15.5%). The interspecific genetic distances of the long 18S sequences ranged from 2.8% between T. apogeana n. sp. and T. paparinii n. sp. to 6.9% between T. apogeana n. sp. and T. palmeri n. sp. (Additional file 4: Table S4). The long 18S sequence of T. apogeana n. sp. (MG758116) was most similar (96.5%) to Theileria sp. (JQ682879) isolated from a burrowing bettong (Bettongia lesueur Quoy and Gaimard) in Western Australia (WA), and T. paparinii n. sp. (MG758115) was most similar (98.9%) to the same Theileria sp. isolate from B. lesueur (JQ682879) (Additional file 2: Table S2). Theileria palmeri n. sp. (MG758113 and MG758120) was most similar (95.8%) to Theileria sp. (MF576261) isolated from Ixodes australiensis Neumann in Australia, and T. worthingtonorum n. sp. (MG758114 and MG758121) was most similar (98.3%) to the same Theileria sp. isolate from I. australiensis (MF576261) (Additional file 2: Table S2).
The long 18S sequences for the novel piroplasms (~1450 bp) and Hepatozoon species (~1650 bp), as well as the short 18S sequences obtained for novel species, are summarised in Table 4.
Novel cf. Sarcocystidae gen. sp.
The short 18S cf. Sarcocystidae Poche, 1913 gen. sp. sequence detected in an I. tasmani tick from a dog in NSW (sample ITF8; MG758127) (100%; 1/1; 95% CI, 2.5–100%) was most similar (98.4%) to species from several different genera in the family Sarcocystidae, including Besnoitia darlingi Mandour, 1965 (MF872603), Toxoplasma gondii Nicolle and Manceaux, 1908 (XR_001974356), Hammondia heydorni Dubey, 1977 (KT184370) and Eimeriidae Minchin, 1903 sp. (KJ634019). Amplification was not obtained for the cf. Sarcocystidae gen. sp. positive sample with the nested PCR, or the HEMO1/HEMO2 PCR, but a 572 bp product was amplified with the HepF300/Hep900 primers. Unexpectedly, a 572 bp cf. Sarcocystidae gen. sp. sequence was obtained (MG758128) and was most similar (96.5%) to Besnoitia Henry, 1913 spp. in GenBank; the top BLAST match was for Besnoitia besnoiti Henry, 1913 (KJ746531) isolated from cattle (Bos taurus Linnaeus) in Croatia (Additional file 2: Table S2).
Genetic distance estimates
The pairwise percent identities of the novel species to the most closely related unnamed species in GenBank ranged from 95.8% between T. palmeri n. sp. (MG758112) and Theileria sp. (MF576261) to 100% between B. lohae n. sp. (MG593272) and Babesia sp. (MG251436) (Table 5). The pairwise identities between the novel and described species ranged from 92.1% between T. palmeri n. sp. (MG758112) and Theileria bicornis Nijhof, 2003 (AF499604) to 98.3% between B. mackerrasorum n. sp. (MG593271) and B. macropus (JQ437265). The average pairwise identities for the novel Babesia, Hepatozoon and Theileria species to 18S sequences from named species in GenBank were 97.4% (standard deviation (SD) 1.3%), 96.1% (SD 0.62%), and 92.5% (SD 0.5%), respectively. The pairwise identities were higher between the two most closely related named species compared to the pairwise identities between the novel species and most closely related named species in all cases, except for B. mackerrasorum n. sp.; the average was 98.3% (SD 1.35%), 96.9% (SD 0.02%) and 94.5% (SD 1.80%) for Babesia, Hepatozoon and Theileria species, respectively. The average pairwise identities for the longer alignments were 98.5% (SD 0.85%), 97.0% (SD 0%) and 94.4% (SD 1.93%) for Babesia, Hepatozoon and Theileria species, respectively. The percentage differences between the pairwise similarities of the shorter versus longer alignments were low, with pairwise identities being overestimated by a maximum of 0.3% and underestimated by a maximum of 0.6% in the shorter alignment (Table 5).
Phylogenetic analyses
The piroplasm phylogenetic tree of the novel species from the present study and named piroplasm species (Fig. 1) shows that T. apogeana n. sp., T. palmeri n. sp., T. paparinii n. sp. and T. worthingtonorum n. sp. grouped within a clade of Theileria species isolated from Australian marsupials with high support (posterior probabilities (pp) ≥ 0.72). Babesia lohae n. sp. and B. mackerrasorum n. sp. both grouped within the Babesia sensu stricto (s.s.) group and formed a clade with B. macropus (Fig. 1). When unnamed species with ≥ 95% were included in the phylogenetic tree and the length of the alignment was increased from 574 bp to 1720 bp to improve taxonomic resolution (Fig. 2), the novel Theileria species still grouped within a clade of sequences isolated from marsupials, but the clade became monophyletic with high support (pp = 1). Within this marsupial clade, T. worthingtonorum n. sp. formed a clade with Theileria sp. (MF576261) isolated from I. australiensis and T. paparinii n. sp. formed a clade with Theileria sp. (JQ682879) isolated from B. lesueur, while T. palmeri n. sp. and T. apogeana n. sp. did not group with any other sequences in the marsupial clade. Babesia lohae n. sp. grouped with other Babesia sp. sequences isolated from I. tasmani (MG251435 and MG251436) (pp ≥ 0.99), while B. mackerrasorum n. sp. grouped closest to, but distinct from, B. macropus isolates from M. giganteus in NSW and QLD (JQ437265 and JQ437266) (pp ≥ 0.55) (Fig. 2).
Bayesian phylogenetic tree of a 574 bp alignment of 18S sequences of named piroplasm species and novel piroplasm sequences derived from this study. The tree was built using the following parameters: HKY85+G+I model; 1,100,000 Markov chain Monte Carlo (MCMC) length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Plasmodium falciparum (JQ627152) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
Bayesian phylogenetic tree of a 1720 bp alignment of 18S sequences of named piroplasm species and novel piroplasm sequences derived from this study, with unnamed sequences with ≥ 95% similarity to novel species included. The tree was built using the following parameters: GTR+G+I model; 1,100,000 MCMC length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Plasmodium falciparum (JQ627152) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
The Adeleorina Léger, 1911 phylogenetic tree constructed from a 406 bp alignment that included novel Hepatozoon species from the present study and described Adeleorina species (Fig. 3) showed that H. banethi n. sp. and H. ewingi n. sp. grouped with other Hepatozoon species with high support (pp = 0.99) and also formed their own distinct clades. When closely related sequences (≥ 96% similar) were included in the phylogenetic reconstruction with a longer alignment length (1457 bp) (Fig. 4), H. banethi n. sp. and H. ewingi n. sp. formed a monophyletic clade with Hepatozoon sp. sequences isolated from D. gliroides (FJ719813 and FJ719814) (pp = 0.62).
Bayesian phylogenetic tree of a 406 bp alignment of 18S sequences of named Adeleorina species and novel Hepatozoon sequences derived from this study. The tree was built using the following parameters: GTR+G+I model; 1,100,000 MCMC length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Cryptosporidium serpentis (AF151376) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
Bayesian phylogenetic tree of a 1457 bp alignment of 18S sequences of named Adeleorina species and novel Hepatozoon sequences derived from this study, with unnamed sequences with ≥ 96% similarity to novel species included. The tree was built using the following parameters: GTR+G+I model; 1,100,000 MCMC length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Cryptosporidium serpentis (AF151376) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
The phylogenetic tree of the cf. Sarcocystidae gen. sp. sequence from the present study and named Sarcocystidae species shows the cf. Sarcocystidae gen. sp. sequence was distinct to all other members of the Sarcocystidae family with low to moderate support (pp = 0.7) (Fig. 5). The phylogenetic tree of cf. Sarcocystidae gen. sp. and members of Eucoccidiorida families showed that cf. Sarcocystidae gen. sp. grouped with the Sarcocystidae family clade with Toxoplasmatinae spp. (pp = 0.52) (Fig. 6).
Bayesian phylogenetic tree of a 629 bp alignment of 18S sequences of named Sarcocystidae species, with fewer Sarcocystis spp. included and the novel cf. Sarcocystidae sp. sequence derived from this study. The tree was built using the following parameters: GTR+G+I model; 1,100,000 MCMC length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Eimeria necatrix (KT184349) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
Bayesian phylogenetic tree of a 668 bp alignment of 18S sequences of Eucoccidiorida families and the novel cf. Sarcocystidae sp. sequence. The tree was built using the following parameters: HKY85+G+I model; 1,100,000 MCMC length; ‘burn-in’ length of 10,000; subsampling frequency of 200. The tree was rooted with the outgroup sequence Babesia rodhaini (AB049999) (not shown). Scale-bar indicates the number of nucleotide substitutions per site
Species descriptions
-
Order Piroplasmida Wenyon, 1926
-
Suborder Piroplasmorina Levine, 1971
-
Family Babesiidae Poche, 1913
-
Genus Babesia Starcovici, 1893
Babesia lohae n. sp.
-
Type-host: Ixodes holocyclus Neumann (Acari: Ixodidae).
-
Type-locality: Park Ridge, Queensland, Australia.
-
Type-material: Bisected tick, tissue extractions and genomic DNA were deposited at the Queensland Museum, Brisbane, Australia, under the accession numbers QMS108579, A015180 and A015181.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG593273 (299 bp 18S rRNA gene) and MG593272 (1430 bp 18S rRNA gene).
-
Vector: The vector potential of I. holocyclus for B. lohae n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Babesia lohae n. sp. is urn:lsid:zoobank.org:act:7526D345-A3A5-4483-8AB6-599F76F0DA32.
-
Etymology: This species is named for Ms Siew-May Loh who discovered Babesia lohae n. sp. in a separate study at the same time as the authors of the present study.
Diagnosis
This organism is a species of Babesia (s.s.) that is genetically distinct from other described Babesia species and forms a clade with other Babesia (s.s.) species isolated from Australian marsupials and ticks (see above).
Babesia mackerrasorum n. sp.
-
Type-host: cf. Haemaphysalis sp. Koch (Acari: Ixodidae).
-
Type-locality: Tanja, New South Wales, Australia.
-
Type-material: Tissue extractions and genomic DNA were deposited at the Australian Museum, Sydney, Australia, under the accession numbers KS.128103.001 and KS.128103.002.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG593276 (299 bp 18S rRNA gene) and MG593271 (1431 bp 18S rRNA gene).
-
Vector: The vector potential of cf. Haemaphysalis sp. for B. mackerrasorum n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Babesia mackerrasorum n. sp. is urn:lsid:zoobank.org:act:BDEDBF3F-28B0-4A4B-B923-5A9EE47EF5B6.
-
Etymology: This species is named after Dr Ian Murray Mackerras (1898–1980) and Dr Mabel Josephine Mackerras (1896–1971) in recognition of their contributions to Australian parasitology.
Diagnosis
This organism is a species of Babesia (s.s.) that is genetically distinct from other described Babesia species and forms a clade with other Babesia (s.s.) species isolated from Australian marsupials and ticks (see above).
-
Order Eucoccidiorida Léger & Duboscq, 1910
-
Suborder Adeleorina Léger, 1911
-
Family Hepatozoidae Wenyon, 1926
-
Genus Hepatozoon Miller, 1908
Hepatozoon banethi n. sp.
-
Type-host: Ixodes tasmani Neumann (Acari: Ixodidae).
-
Type-locality: Devonport, Tasmania, Australia.
-
Other locality: Port Sorell, Tasmania, Australia.
-
Type-material: Bisected tick, tissue extractions and genomic DNA were deposited at the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession numbers K4633 and K4637.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG758134 (303 bp 18S rRNA gene), MG758135 (303 bp 18S rRNA gene), MG758138 (303 bp 18S rRNA gene), MG758133 (1656 bp 18S rRNA gene), MG758135 (1668 bp 18S rRNA gene), and MG758137 (1679 bp 18S rRNA gene).
-
Vector: The vector potential of I. tasmani for H. banethi n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Hepatozoon banethi n. sp. is urn:lsid:zoobank.org:act:B9AC9422-FB14-4BEA-B82C-7A7C901328EA.
-
Etymology: This species is named for Professor Gad Baneth in recognition of his contributions to the field of vector-borne diseases, especially Hepatozoon infections in dogs.
Diagnosis
This organism is a species of Hepatozoon that is genetically distinct from other described Hepatozoon species and forms a clade with other Hepatozoon species isolated from marsupials and ticks (see above).
Hepatozoon ewingi n. sp.
-
Type-host: Haemaphysalis bancrofti Nuttall and Warburton (Acari: Ixodidae).
-
Type-locality: Eungai Creek, New South Wales, Australia.
-
Type-material: Bisected tick, tissue extractions and genomic DNA were deposited at the Australian Museum, Sydney, Australia, under the accession numbers KS.128102.001-KS.128102.003.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG593274 (303 bp 18S rRNA gene) and MG593275 (1680 bp 18S rRNA gene).
-
Vector: The vector potential of H. bancrofti for H. ewingi n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Hepatozoon ewingi n. sp. is urn:lsid:zoobank.org:act:4B0C5B4D-270F-4F3B-8160-1D8F8FA7CA3B.
-
Etymology: This species is named after Professor Sidney Alton Ewing (1934–2018) who contributed more than fifty years of teaching and research to the field of veterinary parasitology.
Diagnosis
This organism is a species of Hepatozoon that is genetically distinct from other described Hepatozoon species and forms a clade with other Hepatozoon species isolated from marsupials and ticks (see above).
-
Order Piroplasmida Wenyon, 1926
-
Suborder Piroplasmorina Levine, 1971
-
Family Theileriidae du Toit, 1918
-
Genus Theileria Bettencourt, Franca & Borges, 1907
Theileria apogeana n. sp.
-
Type-host: Ixodes tasmani Neumann (Acari: Ixodidae).
-
Type-locality: Devonport, Tasmania, Australia.
-
Type-material: Bisected tick, tissue extractions and genomic DNA were deposited at the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession number K4639.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG758126 (790 bp 18S rRNA gene) and MG758116 (1480 bp 18S rRNA gene).
-
Vector: The vector potential of I. tasmani for T. apogeana n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Theileria apogeana n. sp. is urn:lsid:zoobank.org:act:808CAD4C-D259-40E3-B929-E308D23AADBD.
-
Etymology: This species name is derived from the English adjective apogean which relates to the farthest or most distant point.
Diagnosis
This organism is a species of Theileria Bettencourt that is genetically distinct from other described Theileria species and forms a clade with other Theileria species isolated from Australian marsupials and ticks (see above).
Theileria palmeri n. sp.
-
Type-host: Ixodes tasmani Neumann (Acari: Ixodidae).
-
Type-locality: Port Sorell, Tasmania, Australia.
-
Other locality: Devonport, Tasmania, Australia.
-
Type-material: Tissue extractions and genomic DNA were deposited at the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession numbers K4632 and K4638.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG758125 (802 bp 18S rRNA gene), MG758120 (1452 bp 18S rRNA gene), and MG758113 (1506 bp 18S rRNA gene).
-
Vector: The vector potential of I. tasmani for T. palmeri n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Theileria palmeri n. sp. is urn:lsid:zoobank.org:act:6E82C4F6-D069-481F-9752-ADD852F42C57.
-
Etymology: This species is named for Dr Dieter Palmer in recognition of his contributions to the field of parasitology.
Diagnosis
This organism is a species of Theileria that is genetically distinct from other described Theileria species and forms a clade with other Theileria species isolated from Australian marsupials and ticks (see above).
Theileria paparinii n. sp.
-
Type-host: Ixodes tasmani Neumann (Acari: Ixodidae).
-
Type-locality: Lower Wilmot, Tasmania, Australia.
-
Other locality: Devonport, Tasmania, Australia.
-
Type-material: Bisected tick, tissue extractions and genomic DNA were deposited at the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession numbers K4631 and K4635.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG758112 (309 bp 18S rRNA gene), MG758117 (309 bp 18S rRNA gene) and MG758115 (1496 bp 18S rRNA gene).
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Theileria paparinii n. sp. is urn:lsid:zoobank.org:act:0BD6DD5B-5453-416E-8E81-3EEA38DB5FCE.
-
Etymology: This species is named for Dr Andrea Paparini, Murdoch University, Australia, in recognition of his contributions to Australian piroplasm research.
Diagnosis
This organism is a species of Theileria that is genetically distinct from other described Theileria species and forms a clade with other Theileria species isolated from Australian marsupials and ticks (see above).
Theileria worthingtonorum n. sp.
-
Type-host: Ixodes tasmani Neumann (Acari: Ixodidae).
-
Type-locality: Port Sorell, Tasmania, Australia.
-
Type-material: Tissue extractions and genomic DNA were deposited at the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession numbers K4634 and K4636.
-
Representative DNA sequences: DNA sequences were deposited in GenBank under the accessions MG758118 (310 bp 18S rRNA gene), MG758119 (310 bp 18S rRNA gene), MG758121 (1497 bp 18S rRNA gene) and MG758114 (1504 bp 18S rRNA gene).
-
Vector: The vector potential of I. tasmani for T. worthingtonorum n. sp. is unknown.
-
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [24], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D1B6E4E1-168C-488B-B809-381642900749. The LSID for the new name Theileria worthingtonorum n. sp. is urn:lsid:zoobank.org:act:89A747A5-5D91-47A1-84EB-AF01236145B0.
-
Etymology: This species is named for the first author’s grandparents Mr Peter Ross Worthington and Mrs Dawn Rose Worthington.
Diagnosis
This organism is a species of Theileria that is genetically distinct from other described Theileria species and forms a clade with other Theileria species isolated from Australian marsupials and ticks (see above).
Discussion
This study is the first to investigate, on a national scale, apicomplexan parasites carried by ticks that parasitise companion animals in Australia. Although we hypothesised that species of endemic piroplasms would be detected in ticks parasitising dogs, cats and horses, the discovery of nine novel apicomplexan species was unexpected. The low pairwise identities and distinct phylogenetic groupings of the novel 18S sequences to the most closely related described species supports the new species classifications. This investigation used an approach similar to that used by Schnittger et al. [25] to taxonomically assign the sequences to species. All Babesia and Theileria species sequenced in this study were ≤ 96.4% and ≤ 93.3% similar to the most closely related named species, respectively, with the exception of B. mackerrasorum, which was 98.3% similar to B. macropus (JQ437265). The highest pairwise similarity for the novel Hepatozoon species compared to named Hepatozoon species was 96.6%, which is less than the pairwise identity of the recently described Hepatozoon musa Borges-Nojosa, 2017 to its most closely related described species (~99% similar) [26].
Moreover, the new species assignments are further supported by the distinct phylogenetic groupings of the novel species to named species (Figs. 1 and 3). Babesia mackerrasorum n. sp. and B. lohae n. sp. are most closely related to Babesia spp. that have been isolated previously from Australian marsupials and ticks from marsupials (Figs. 1 and 2). Babesia mackerrasorum n. sp. clustered separately with a longer branch length to B. macropus (JQ437265 and JQ437266) isolated from eastern grey kangaroos (M. giganteus) in NSW and QLD. Additionally, the intraspecific genetic variation of B. macropus at the 18S gene is 0.2% [27, 28], and as the interspecific genetic variation at the 18S gene between B. macropus and B. mackerrasorum n. sp. was 1.7%, this also suggests that they are different species. Babesia lohae n. sp. is 100% similar to an unnamed Babesia sp. sequence (MG251436) derived from a common marsupial tick (I. tasmani), collected from a common brushtail possum (T. vulpecula), also in QLD (unpublished), and thus it is possible that the brushtail possum is a native reservoir host of B. lohae n. sp., and although a number of Babesia species have been identified in the blood of native marsupials [27,28,29,30], no studies have yet investigated brushtail possums for Babesia spp.
The novel Theileria species also phylogenetically clustered with 18S sequences derived from native marsupials (Figs. 1 and 2), suggesting that the Theileria species identified in I. tasmani have marsupial hosts. Theileria paparinii n. sp. is most closely related to, but distinct from, the previously described Theileria penicillata Clark and Spencer 2007 and Theileria brachyuri Clark and Spencer 2007 that were isolated from woylies and quokkas, respectively, in WA [31, 32]. Theileria worthingtonorum n. sp. is most closely related to Theileria fuliginosa Clark and Spencer 2007 from the western grey kangaroo (Macropus fuliginosus Desmarest) in WA [31]. Ixodes tasmani has been described as catholic in its feeding habits having been recorded on 42 host species, including marsupials, monotremes, rodents and domestic animals [8]. It is likely that there are multiple native host species for the novel Theileria species, and for H. banethi n. sp. that was also identified in I. tasmani. Both H. ewingi n. sp. (isolated from H. bancrofti, wallaby tick, which feeds on bandicoots, possums, macropods and other marsupials [9]) and H. banethi n. sp. group with Hepatozoon spp. sequenced from D. gliroides [33], a marsupial from Chile. This again lends weight to the suggestion that these new Australian Hepatozoon species have native marsupial hosts. Other Hepatozoon species have been described from Australian wildlife, including reptiles [34,35,36,37], ticks [38] and bandicoots (Perameles Geoffroy Saint-Hilaire spp. and Isoodon Desmarest spp.) [37], the latter of which was < 97% similar to the Hepatozoon species from this study (data not shown).
Although many apicomplexan species are difficult to morphologically differentiate between or are indistinguishable at the species level [38], there are distinct morphological differences between families and genera. Therefore, morphological characterisations, with additional genetic characterisations, are required to confirm the family and genus of cf. Sarcocystidae gen. sp., although based on the pairwise distances and phylogenetic reconstructions, this sequence certainly represents a novel species.
This study has demonstrated that the use of conventional PCR and Sanger sequencing for characterising apicomplexans in ticks is limited due to the identification of co-infections of piroplasms and Hepatozoon species, and co-infections of multiple Theileria species (outlined in Table 4). To more comprehensively identify co-infections in ticks, a next-generation sequencing (NGS) approach could be used, which has been shown to be a useful technique for identifying Trypanosoma Gruby, 1843 spp. in ticks [39]. There are likely other protozoans in native ticks that remain to be discovered, and future studies could aim to extend this NGS approach to protozoans in general, which would greatly improve the speed and lower the cost for studies that aim to broadly screen for protists.
The prevalence of novel apicomplexan species detected in this study was relatively high (1.3%; 9/711; 95% CI: 0.6–2.4%), which reflects that apicomplexan species in ticks and their hosts in Australia has been greatly understudied to date. Previous studies that have investigated the prevalence of piroplasms and Hepatozoon spp. in Australian wildlife have reported these Apicomplexa to be highly prevalent in their hosts. For example, T. penicillata has been reported in Bettongia penicillata Gray in WA at a prevalence of 80.4% (123/153) [32]. Hepatozoon sp. has been found in 34.1% (15/44) of I. tasmani ticks collected from Tasmanian devils (Sarcophilus harrisii Boitard) [40], while the prevalence of H. banethi n. sp. in I. tasmani ticks tested in this study was considerably lower (5.1%; 3/59; 95% CI: 1.1–14.1%). Hepatozoon sp. has been detected in the southern brown bandicoot (Isoodon obesulus Shaw) in WA in 58.1% (18/31) of samples [37], and a high prevalence of Hepatozoon spp. has also been reported in reptiles. For example, a study by Jakes et al. [34] detected Hepatozoon boigae Mackerras, 1961 in 29% of 97 brown tree snakes (Boiga irregularis Merrem), and Hepatozoon spp. has been detected in 35.6% of 35 blood samples from water pythons (Liasis fuscus Peters) with 57.7% of 187 ticks collected from L. fuscus also positive for Hepatozoon spp. [38], and another study has detected Hepatozoon spp. in 100% of 100 blood samples from L. fuscus [16].
It is not surprising that B. vogeli and T. orientalis genotype 2 (Ikeda) were identified in R. sanguineus and H. longicornis, respectively. Rhipicephalus sanguineus is a competent vector of B. vogeli [5, 6] and H. longicornis is likely a vector of T. orientalis genotype Ikeda [41]. The finding of H. canis, a tick-borne pathogen of dogs that, to our knowledge, has not been recorded in Australia previously, was unanticipated. Of note is that H. canis was detected in a paralysis tick, I. holocyclus, which has not been associated with H. canis before. Studies have shown that R. sanguineus is a vector [42] and H. longicornis is a putative vector [43] of H. canis. These species are present in Australia, and although no R. sanguineus (0/183; 95% CI: 0–2%) or H. longicornis (0/75; 95% CI: 0–4.8%) ticks examined in the present study contained H. canis DNA, the prevalence of H. canis in ticks collected from dogs in endemic areas has been reported to be as low as 1.5% (4/267) [44]. It is also possible that the I. holocyclus tick (which was engorged with the host’s blood) ingested a H. canis-infected blood meal from the host. This is not the first instance of an exotic tick-borne pathogen of companion animals in Australia; in the 1970s, there was a brief incursion of Theileria equi in imported horses, which caused localised outbreaks of equine piroplasmosis [45,46,47]. Without this broad investigation for piroplasms and Hepatozoon spp. in ticks, it is likely that H. canis would have remained undetected, which emphasises the need for ongoing surveillance of tick-borne pathogens across the country. Australian biosecurity authorities have been advised of this finding and an investigation into the potential sources and dissemination of this presumed incursion of H. canis is under way.
Conclusion
This investigation of more than 700 ticks in Australia has led to the discovery of nine new apicomplexans, the exotic canine pathogen H. canis, and detected the endemic tick-borne pathogens B. vogeli and T. orientalis genotype Ikeda. Future studies are required to establish the host range and the vector competence of ticks for the newly described apicomplexans as these haemoprotozoans could represent an infectious disease threat to companion animal species.
Abbreviations
- 18S :
-
18S ribosomal RNA gene
- BIC:
-
Bayesian Information Criterion
- ExCs:
-
Extraction reagent blank controls
- MPSP:
-
Major piroplasm surface protein
- nr/nt:
-
Non-redundant nucleotide
- NSW:
-
New South Wales
- NT:
-
Northern Territory
- NTCs:
-
No-template controls
- pp:
-
Posterior probabilities
- QLD:
-
Queensland
- SA:
-
South Australia
- SD:
-
Standard deviation
- SNPs:
-
Single nucleotide polymorphisms
- Tann:
-
Annealing temperature
- TAS:
-
Tasmania
- VIC:
-
Victoria
- WA:
-
Western Australia
References
Day MJ. One health: the importance of companion animal vector-borne diseases. Parasit Vectors. 2011;4:49.
Jefferies R, Ryan UM, Muhlnickel CJ, Irwin PJ. Two species of canine Babesia in Australia: detection and characterization by PCR. J Parasitol. 2003;89:409–12.
Brown GK, Canfield PJ, Dunstan RH, Roberts TK, Martin AR, Brown CS, et al. Detection of Anaplasma platys and Babesia canis vogeli and their impact on platelet numbers in free-roaming dogs associated with remote Aboriginal communities in Australia. Aust Vet J. 2006;84:321–5.
Muhlnickel CJ, Jefferies R, Morgan-Ryan UM, Irwin PJ. Babesia gibsoni infection in three dogs in Victoria. Aust Vet J. 2002;80(10):606.
Hill MWM, Bolton BL. Canine Babesiosis in Queensland. Aust Vet J. 1966;42:391–2.
Irwin PJ, Hutchinson GW. Clinical and pathological findings of Babesia infection in dogs. Aust Vet J. 1991;68:204–9.
Greay TL, Oskam CL, Gofton AW, Rees RL, Ryan UM, Irwin PJ. A survey of ticks (Acari: Ixodidae) of companion animals in Australia. Parasit Vectors. 2016;9:1–10.
Barker SC, Walker AR. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa. 2014;3816:1–144.
Roberts FHS. Australian ticks. Melbourne: Commonwealth Scientific and Industrial Research Organization; 1970.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Jefferies R, Ryan UM, Irwin PJ. PCR-RFLP for the detection and differentiation of the canine piroplasm species and its use with filter paper-based technologies. Vet Parasitol. 2007;144:20–7.
Paparini A, Ryan UM, Warren K, McInnes LM, de Tores P, Irwin PJ. Identification of novel Babesia and Theileria genotypes in the endangered marsupials, the woylie (Bettongia penicillata ogilbyi) and boodie (Bettongia lesueur). Exp Parasitol. 2012;131:25–30.
Oosthuizen MC, Zweygarth E, Collins NE, Troskie M, Penzhorn BL. Identification of a novel Babesia sp. from a sable antelope (Hippotragus niger Harris, 1838). J Clin Microbiol. 2008;46:2247–51.
Matjila PT, Leisewitz AL, Oosthuizen MC, Jongejan F, Penzhorn BL. Detection of a Theileria species in dogs in South Africa. Vet Parasitol. 2008;157:34–40.
Zakimi S, Kim JY, Oshiro M, Hayashida K, Fujisaki K, Sugimoto C. Genetic diversity of benign Theileria parasites of cattle in the Okinawa Prefecture. J Vet Med Sci. 2006;68:1335–8.
Ujvari B, Madsen T, Olsson M. High prevalence of Hepatozoon spp. (Apicomplexa, Hepatozoidae) infection in water pythons (Liasis fuscus) from tropical Australia. J Parasitol. 2004;90:670–2.
Perkins SL, Keller AK. Phylogeny of nuclear small subunit rRNA genes of hemogregarines amplified with specific primers. J Parasitol. 2001;87:870–6.
Criado-Fornelio A, Ruas JL, Casado N, Farias NA, Soares MP, Muller G, et al. New molecular data on mammalian Hepatozoon species (Apicomplexa: Adeleorina) from Brazil and Spain. J Parasitol. 2006;92:93–9.
Tanaka M, Onoe S, Matsuba T, Katayama S, Yamanaka M, Yonemichi H, et al. Detection of Theileria sergenti infection in cattle by polymerase chain reaction amplification of parasite-specific DNA. J Clin Microbiol. 1993;31:2565–9.
Yang R, Murphy C, Song Y, Ng-Hublin J, Estcourt A, Hijjawi N, et al. Specific and quantitative detection and identification of Cryptosporidium hominis and C. parvum in clinical and environmental samples. Exp Parasitol. 2013;135:142–7.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5.
ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–9.
Schnittger L, Yin H, Gubbels MJ, Beyer D, Niemann S, Jongejan F, et al. Phylogeny of sheep and goat Theileria and Babesia parasites. Parasitol Res. 2003;91:398–406.
Borges-Nojosa DM, Borges-Leite MJ, Maia JP, Zanchi-Silva D, da Rocha Braga R, Harris DJ. A new species of Hepatozoon Miller, 1908 (Apicomplexa: Adelerina) from the snake Philodryas nattereri Steindachner (Squamata: Dipsadidae) in northeastern Brazil. Syst Parasitol. 2017;94:65–72.
Dawood KE, Morgan JAT, Busfield F, Srivastava M, Fletcher TI, Sambono J, et al. Observation of a novel Babesia spp. in eastern grey kangaroos (Macropus giganteus) in Australia. Int J Parasitol Parasites Wildl. 2013;2:54–61.
Donahoe SL, Peacock CS, Choo AYL, Cook RW, O'Donoghue P, Crameri S, et al. A retrospective study of Babesia macropus associated with morbidity and mortality in eastern grey kangaroos (Macropus giganteus) and agile wallabies (Macropus agilis). Int J Parasitol Parasites Wildl. 2015;4:268–76.
Backhouse TC, Bolliger A. Babesia tachyglossi n. sp. from the echidna Tachyglossus aculeatus. J Protozool. 1959;6:320–2.
Mackerras M. The haematozoa of Australian mammals. Aust J Zool. 1959;7:105–35.
Clark P, Spencer PBS. Description of three new species of Theileria Bettencourt, Franca & Borges, 1907 from Macropodoidea in Western Australia. Trans R Soc S Aust. 2007;131:100–6.
Rong J, Bunce M, Wayne A, Pacioni C, Ryan U, Irwin P. A high prevalence of Theileria penicillata in woylies (Bettongia penicillata). Exp Parasitol. 2012;131:157–61.
Merino S, Vásquez RA, Martínez J, Celis-Diez JL, Gutiérrez-Jiménez L, Ippi S, et al. Molecular characterization of an ancient Hepatozoon species parasitizing the ‘living fossil’ marsupial ‘Monito del Monte’ Dromiciops gliroides from Chile. Biol J Linn Soc. 2009;98:568–76.
Jakes KA, O'Donoghue PJ, Whittier J. Ultrastructure of Hepatozoon boigae (Mackerras, 1961) nov. comb. from brown tree snakes, Boiga irregularis, from northern Australia. Parasitol Res. 2003;90:225–31.
Jakes K, O'Donoghue PJ, Cameron SL. Phylogenetic relationships of Hepatozoon (Haemogregarina) boigae, Hepatozoon sp., Haemogregarina clelandi and Haemoproteus chelodina from Australian reptiles to other Apicomplexa based on cladistic analyses of ultrastructural and life-cycle characters. Parasitology. 2003;126:555–9.
O'Donoghue P, Adlard R. Catalogue of protozoan parasites recorded in Australia. Mem Queensl Mus. 2000;45:1–163.
Wicks RM, Spencer PB, Moolhuijzen P, Clark P. Morphological and molecular characteristics of a species of Hepatozoon Miller, 1908 (Apicomplexa: Adeleorina) from the blood of Isoodon obesulus (Marsupialia: Peramelidae) in Western Australia. Syst Parasitol. 2006;65:19–25.
Vilcins IM, Ujvari B, Old JM, Deane E. Molecular and morphological description of a Hepatozoon species in reptiles and their ticks in the Northern Territory, Australia. J Parasitol. 2009;95:434–42.
Barbosa AD, Gofton AW, Paparini A, Codello A, Greay T, Gillett A, et al. Increased genetic diversity and prevalence of co-infection with Trypanosoma spp. in koalas (Phascolarctos cinereus) and their ticks identified using next-generation sequencing (NGS). PLoS One. 2017;12:e0181279.
Vilcins I-ME, Old JM, Deane E. Detection of a Hepatozoon and spotted fever group Rickettsia species in the common marsupial tick (Ixodes tasmani) collected from wild Tasmanian devils (Sarcophilus harrisii). Tasmania. Vet Parasitol. 2009;162:23–31.
Hammer JF, Emery D, Bogema DR, Jenkins C. Detection of Theileria orientalis genotypes in Haemaphysalis longicornis ticks from southern Australia. Parasit Vectors. 2015;8:229.
Baneth G, Samish M, Shkap V. Life cycle of Hepatozoon canis (Apicomplexa: Adeleorina: Hepatozoidae) in the tick Rhipicephalus sanguineus and domestic dog (Canis familiaris). J Parasitol. 2007;93:283–99.
Murata T, Inoue M, Taura Y, Nakama S, Abe H, Fujisaki K. Detection of Hepatozoon canis oocyst from ticks collected from the infected dogs. J Vet Med Sci. 1995;57:111–2.
Dantas-Torres F, Latrofa MS, Weigl S, Tarallo VD, Lia RP, Otranto D. Hepatozoon canis infection in ticks during spring and summer in Italy. Parasitol Res. 2012;110:695–8.
Churchill RC, Best DR. Babesiosis of a horse in Australia. Aust Vet J. 1976;52:487.
Mahoney D, Wright I, Frerichs W, Groenendyk S, O'Sullivan B, Roberts M, et al. The identification of Babesia equi in Australia. Aust Vet J. 1977;53:461–4.
Martin R. Equine piroplasmosis: the temporary importation of seropositive horses into Australia. Aust Vet J. 1999;77:308–9.
Rozsa L, Reiczigel J, Majoros G. Quantifying parasites in samples of hosts. J Parasitol. 2000;86:228–32.
Acknowledgements
Publication of this paper has been sponsored by Bayer Animal Health in the framework of the 13th CVBD World Forum Symposium. We thank Ms Frances Brigg from the Western Australian State Agricultural Biotechnology Centre for her assistance with Sanger sequencing, Dr Dieter Palmer from the Department of Primary Industries and Regional Development for assisting with positive controls for the study and Dr Andrea Paparini for allowing us to use his 18SApiF/18SApiR primer set for the study. We also thank The Australian Centre for Wildlife Genomics at the Australian Museum, Dr Jessica Worthington Wilmer and Dr Owen Seeman from the Queensland Museum, Dr Catherine Byrne and Dr Kirrily Moore from the Department of State Growth at the Tasmanian Museum and Art Gallery and Dr Mark Harvey from the Western Australian Museum. Lastly, we acknowledge Bayer Australia area managers and the general public for collecting the ticks.
Funding
This research was funded by the Australian Research Council (Linkage Project 130100050), Bayer Australia Ltd and Bayer AG (Germany). TLG is supported by a Murdoch University Postgraduate Research scholarship.
Availability of data and materials
The data supporting the conclusions of this article are included within the article. The sequences were submitted in the GenBank database under the accession numbers MG062865, MG571580-MG571582, MG593271-MG593276, MG758109-MG758121 and MG758124-MG758138. Type material was submitted to the Australian Museum, Sydney, Australia, under the accession numbers KS.128102 and KS.128103, the Queensland Museum, Brisbane, Australia, under the accession numbers QMS108579, A015180 and A015181 and the Tasmanian Museum and Art Gallery, Hobart, Australia, under the accession numbers K463-K4640.
Author information
Authors and Affiliations
Contributions
TLG contributed in the design of the study, performed tick identification, DNA extractions, PCRs, Sanger sequencing preparation and phylogenetic analyses and wrote the manuscript. AZ performed PCRs, gel electrophoresis and PCR purifications and contributed to the preparation of the manuscript. A-SK and JMO performed PCRs and contributed to the preparation of the manuscript. RLR contributed to conceiving, designing and coordinating the study, tick collection and preparation of the manuscript. UMR contributed to the design of the study and preparation of the manuscript. CLO performed PCRs and contributed to the design of the study and preparation of the manuscript. PJI conceived, designed and coordinated the study and contributed to preparation of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Collection locations of ticks from dogs, cats and horses that were screened for piroplasms and Hepatozoon spp. (PDF 261 kb)
Additional file 2:
Table S2. Summary of PCR screening and NCBI BLAST results for apicomplexan sequences obtained in this study. (PDF 200 kb)
Additional file 3:
Table S3. Prevalence of Apicomplexa species Australia-wide and in each state and territory summarised for all tick species from all hosts, individual tick species from all hosts, and individual tick species from dogs, cats and horses. (XLSX 35 kb)
Additional file 4:
Table S4. Pairwise genetic distance matrix of 18S sequences similarities (%) from novel piroplasms and Hepatozoon spp. described in this study. (PDF 202 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Greay, T.L., Zahedi, A., Krige, AS. et al. Endemic, exotic and novel apicomplexan parasites detected during a national study of ticks from companion animals in Australia. Parasites Vectors 11, 197 (2018). https://doi.org/10.1186/s13071-018-2775-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2775-y