Abstract
Background
Theileria are blood-borne intracellular protozoal parasites belonging to the phylum Apicomplexa. Previously considered a benign parasite in Australia, outbreaks of clinical disease resulting from Theileria orientalis genotypes have been reported in Australia since 2006. Since this time, outbreaks have become widespread in south-eastern Australia, resulting in significant adverse impacts on local dairy and beef industries. This paper provides the first investigation into the possible biological and mechanical vectors involved in the rapid spread of the parasite.
Methods
To identify possible vectors for disease, ticks, biting flies and mosquitoes were collected within active outbreak regions of Gippsland, Victoria. Ticks were collected from cattle and wildlife, and mosquitoes and biting flies were collected in traps in close proximity to outbreak herds. Ticks were identified via DNA barcoding of the mitochondrial cytochrome oxidase I gene. Barcoded ticks were pooled according to species or phylogenetic group and tested for the presence of T. orientalis and the genotypes Ikeda, Chitose and Buffeli using real-time PCR.
Results
DNA barcoding and phylogenetic analysis identified ticks from the following species: Haemaphysalis longicornis, Ixodes holocyclus, Ixodes cornuatus, Ixodes hirsti, and Bothriocroton concolor. Additional Haemaphysalis, Ixodes and Bothriocroton spp. were also identified. Of the ticks investigated, only H. longicornis ticks from cattle carried theilerial DNA, with the genotypes Ikeda, Chitose and Buffeli represented. Mosquitoes collected in close proximity to outbreak herds included; Aedes camptorhynchus, Aedes notoscriptus, Coquillettidia linealis, Culex australicus, and Culex molestus. Low levels of T. orientalis Buffeli genotype were detected in some mosquitoes. The haematophagous flies tested negative.
Conclusions
This is the first demonstration of a potential vector for T. orientalis in the current Australasian disease outbreak.
Similar content being viewed by others
Background
Theileria are blood-borne intracellular protozoal parasites belonging to the phylum Apicomplexa. Historically, the precise classification of the various genotypes of the parasite has been the cause of some confusion [1-3]. In particular, the agent of the more benign form of bovine theileriosis was speciated differently, being named as T. buffeli in Australia, T. sergenti in Japan and East Asia, and T. orientalis in many other locations [2]. The name Theileria orientalis is now widely applied to the species present in Australia, New Zealand and throughout Asia [4-15]. It is now appreciated that T. orientalis can be separated into several genotypes, namely, type 1 (Chitose), type 2 (Ikeda), type 3 (Buffeli), types 4–8 [6,7] and types N1-N3 [14].
Theileria has been recognised in Australia since 1910 [16] and is found in all states except Tasmania [8]. T. orientalis has long been considered a benign parasite [4,6,16,17]. However, pathogenic genotypes are now recognised in many countries including Australia [7,10], New Zealand [5], Japan, [11], China [18] and Korea [19]. Since 2006, a large number of outbreaks of clinical disease have been reported in New South Wales, Victoria, Western Australia [7,10,20], and more recently South Australia [21]. The emergence of clinical theileriosis and associated mortalities in Australia is of increasing concern for the local beef and dairy industries [7,8]. Clinical signs of T. orientalis are mostly associated with the sequelae from anaemia. These signs can include lethargy, weakness, anorexia, pale mucous membranes, lymph node swelling, tachypnoea, tachycardia, dyspnoea, jaundice, late-term abortion, dystocia, pyrexia, and mortality [5,7,8].
The method of introduction of Theileria to Australia is uncertain and may have occurred with the introduction of Haemaphysalis longicornis ticks into Australia [16,22,23]. Moreover, the emergence of more pathogenic genotypes remains unclear, although it has been speculated that the Ikeda genotype was introduced to Australia from cattle imported from Eastern Asia [24]. A severe outbreak of theileriosis on a beef farm has been linked to T. orientalis Ikeda and Chitose genotype entering the more southern, temperate area of Victoria [7]. It is known that the Ikeda genotype is more associated with clinical disease than Chitose and Buffeli in Australia, and this also occurs internationally where clinical cases of T. orientalis have been reported [10,11]. Currently, the Australian vectors for T. orientalis Ikeda genotype have not been determined.
Haemaphysalis longicornis, a known vector tick for T. orientalis in other countries [2], has a complex life cycle. It has a wide host range, and a distribution along the coastal strip from south east Queensland to the Victoria border with rare occurrences in other locations in Victoria [25]. It has also been reported in locations in south west Western Australia [26]. It is a three host tick with larva, nymph, and adult engorging for approximately 7 days before dropping from the host [27]. Female ticks lay up to 2000 eggs in late spring and early summer (November to January) which hatch in 60–90 days depending on environmental conditions. All stages can survive more than 200 days without feeding [27]. Most adults are seen in January and February, reproduction is via parthenogenesis, and so male adults are rarely seen. Other ticks recorded from cattle in Victoria include; Ixodes cornuatus, Ixodes holocyclus, Amblyomma moreliae, Bothriocroton concolor (formerly Aponomma concolor), Aponomma fimbriatum, Bothriocroton hydrosauri (formerly Aponomma hydrosauri), and Rhipicephalus sanguineus [25,28]. Of these ticks, I. cornuatus and I. holocyclus are known from Eastern Victoria where this research was conducted [29]. In addition, Amblyomma moreliae has also been recorded in Gippsland [25]. Bothriocroton concolor is mostly seen on echidnas, Bothriocroton hydrosauri is mostly found on reptiles, and Rhipicephalus sanguineus is mostly seen on dogs and the occurrence of all of these ticks on cattle is rare [25]. The role of H. longicornis has been investigated as a vector of “T. buffeli” in Australia. This species was once implicated as a likely vector [30], but was discounted in subsequent studies [1,22]. Attempts to experimentally transmit T. orientalis Warwick strain (Buffeli type) to cattle via the trans-stadial and transovarial routes were unsuccessful while trans-stadial transmission was routinely achieved with H. humerosa [22,31]. Conversely in Japan, experimental transmission studies employing the Japanese Ikeda genotype of T. orientalis with H. longicornis sourced from both Australian and Japanese populations were successful. Indeed, these early studies were used as one criterion to differentiate the Australian “T. buffeli” (now referred to as T. orientalis Buffeli) from “T. sergenti” which encompasses the Ikeda genotype [2]. In separate experiments, the native Australian wallaby tick, H. bancrofti, was shown to transmit both T. orientalis Ikeda [2], and a strain of T. orientalis sourced from Queensland and presumed to be the Buffeli type “Warwick strain” [32]. Both H. bancrofti and H. humerosa are apparently competent vectors for T. orientalis under experimental conditions, but the range of both of these species is principally limited to the more temperate and tropical climates of the northern coast of New South Wales and coastal Queensland. Although H. bancrofti has been recorded sporadically from Victoria [33] this tick and H. humerosa are less likely to act as vectors in Victoria. Taken together, these data suggest that the most likely vectors of T. orientalis might include H. longicornis, H. bancrofti, I. cornuatus, I. holocyclus, and A. moreliae.
Methods
Sample collection
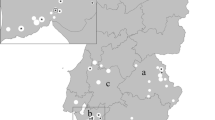
The collection of ticks and arthropods invertebrates described did not require ethics approval. Local producers and wildlife experts provided ticks taken from animals in their care. For this study, 220 ticks were collected from a variety of livestock and wildlife hosts over a large geographical region of Victoria (Figure 1) by local veterinary practices, wildlife carers, farmers and members of the public. Ticks were predominantly of the adult stage, although some larval and nymph stage ticks were also collected, and ranged from unengorged through to fully engorged. Haematophagous flies were collected during suitable weather during summer 2013, autumn 2014, and summer of 2015, in locations within one kilometre of outbreak farms. Two Nzi traps (Rincon-Vitova Insectaries, Ventura, CA, USA) were set up using dry ice as an attractant with 1-Octen-3-ol. A total of 4 (UV) light traps (Australian Entomology Supplies, Model PR101) were used for the collection of mosquitoes. Each trap was baited with carbon dioxide from dry ice. Traps were set on properties where clinical cases of theileriosis had been identified, or located in close proximity not more than 1 kilometre from such a herd. Capture data was recorded for all arthropods, which were then frozen and stored at −80°C until required.
Distribution of Haemaphysalis longicornis in Australia. Map of Australia showing the known range of H. longicornis as described in [30]. An enlarged map of the state of Victoria is also shown with the geographic locations, from which ticks and other arthropods were collected, highlighted (closed circles). The location of the likely entry point of T. orientalis Ikeda into the state of Victoria is also shown (open circle).
Arthropod identification
Mosquitoes were identified morphologically using a dichotomous key [34]. Mosquito species identified included Aedes camptorhynchus, Aedes notoscriptus, Coquillettidia linealis, Culex australicus, and Culex molestus. Preliminary identification of ticks was undertaken using identification charts [25]. All ticks collected were subsequently transported on dry ice to the laboratory for molecular identification. Ticks were photographed, in order to maintain a record of sample morphology, and samples for which morphological identification was uncertain (n = 135), were subjected to DNA barcoding. A H. longicornis tick from an Australian reference collection (AgriBio, Centre for AgriBioscience) was also barcoded for comparison with the field-collected ticks. For PCR testing, an individual leg was dissected from each tick using separate sterile forceps and scalpel blades and the legs were washed with PBS to remove any adherent host material. DNA was extracted from the tick legs using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions and with a 100 μL elution volume. A process control which contained no tissue was included with each DNA extraction run to control for cross-contamination of samples. The mitochondrial cytochrome oxidase I (COI) genes were amplified using the metazoan “barcoding” primers of Folmer [35]. PCR amplification was performed in a total volume of 25 μL comprising 1 × Immolase reaction buffer, 2.0 mM MgCl2, 200 μM dNTPs, 400 nM each of the LCO1490 and HCO2198 primers and 1 U of Immolase DNA polymerase (Bioline). PCR products were visualised on a 0.5 × TBE-1.5% agarose gel and of the 135 ticks tested, 105 yielded sufficient amplicon for downstream sequencing. PCR products were purified using the Qiaquick PCR purification kit (Qiagen) and subjected to Big Dye terminator sequencing at the Australian Genome Research Facility (AGRF). Contiguous sequences were assembled using Geneious Version 7.1.5 (Biomatters, Auckland, New Zealand) and compared with existing tick COI sequences in GenBank using the Basic Local Alignment Search Tool (BLAST).
Detection of T. orientalis by quantitative PCR
Ticks grouped by species or phylogenetic cluster, and where possible by host and location, were dissected for quantitative PCR (qPCR) analysis by removal and collection of the capitulum and the anterior portion of the scutum. Separate sterile forceps and scalpel blades were used for the dissection of each tick. For mosquitoes and march flies, tissue from the head, abdomen and thorax was extracted. Tick tissue was pooled in groups of 1–10 samples depending on tissue volume to yield a total mass of 10–15 mg per pool. March flies (Dasybasis sp.) were tested individually by dissecting them laterally to yield 10–15 mg of tissue. Mosquitoes were also tested in 10–15 mg pools. Thirteen pools of mosquitoes were tested (approximately 10 mosquitoes per pool). All remaining mosquitoes were batch tested. DNA extraction was performed as described above for the tick legs, except that each tissue sample was homogenised prior to extraction with a sterile microfuge pestle. qPCR was performed using a validated multiplex qPCR assay for T. orientalis detection and genotype differentiation as described previously [36]. This assay was used for quantification of the total load of T. orientalis (Universal) and to semi-quantitatively determine the relative proportions of the Ikeda and Chitose genotypes (UIC assay). DNA extracts were also tested for the Buffeli genotype of T. orientalis using a semi-quantitative singleplex qPCR (B assay). For this assay, the same reagents and instrumentation described in Bogema et al. [36] were used, however, the probe mix was substituted for a single probe design based on an in silico analysis to specifically target the Buffeli genotype (5′ - FAM-CTCCTTTGCAGTATTCTTCTATCTC-BHQ1 - 3′). The analytical specificity of the Buffeli probe for this genotype was assessed using plasmids containing Ikeda Chitose or Buffeli MPSP gene inserts and also DNA extracts from T. orientalis-negative cattle and cattle infected with the closely related parasites, Babesia bovis and B. bigemina all as described in Bogema et al. [36]. The limit of detection (LOD) was defined as the limit where 95% of assays were successful and was experimentally determined by testing 8 replicates of Buffeli plasmid DNA at dilutions 15000, 1500, 150, 50, 15, 5, 1.5 and 0.5 gene copies/μL (GC/μL), followed by Probit analysis. The efficacy of the Buffeli singleplex assay was confirmed on bovine blood extracts previously demonstrated to be positive for this genotype [9]. A standard curve generated from a 10-fold dilution series of a plasmid containing the Buffeli MPSP gene [36] ranged from 1.5 × 101 to 1.5 × 107 to MPSP GC/μL and was included in each run. Quality acceptance parameters for standard curves were an R2 value > 0.98 and an amplification efficiency between 90 and 110%.
Results
Identification of tick species
Morphological identification and molecular testing aligned for all specimens examined. A number of ticks were not examined morphologically due to damage to the tick during collection limiting our ability to make an accurate identification. A total of 81 tick COI barcodes were identified; 56 from ticks of the Haemaphysalis genus, 18 from Ixodes spp. and 7 from Bothriocroton spp. A phylogenetic tree constructed from a MUSCLE alignment of these sequences and GenBank reference sequences is shown in Figure 2. Tick barcode sequences clustered into 9 major clades, 8 with high (>97%) bootstrap support. Species identified included H. longicornis, B. concolor, I. holocyclus, I. hirsti and I. cornuatus. The remaining 4 clades identified also clustered within the genera Haemaphysalis, Bothriocroton and Ixodes, however these tick samples could not be confidently assigned to particular species. Furthermore, the four unidentified clades included damaged ticks unsuitable for morphological identification. There was moderate bootstrap support for a relationship between Haemaphysalis ticks collected from koalas (Phascolarctos cinereus victor) and the species H. doenitzi and H. humerosa. Ticks collected from the common wombat (Vombatus ursinus) were of the Bothriocroton genus but formed a distinct cluster from B. concolor (the echidna tick), and B. undatum and B. hydrosauri, which are typically found on reptiles. A further cluster of sequences forming a sister group to I. hirsti was identified from both koalas (P. cinereus victor) and brush-tailed possums (Trichosurus vulpecula). Tick sample 30–1, also collected from T. vulpecula, was most closely related to members of the Ixodes genus, but did not cluster with any other species for which sequence data was available.
Phylogenetic tree of ticks investigated. Phylogenetic tree based on mitochondrial cytochrome oxidase I (COI) gene sequences of ticks collected in this study along with reference tick COI sequences sourced from Genbank. The phylogenetic tree is based on a MUSCLE alignment of the sequences in which gap positions were trimmed and was constructed using the Maximum Likelihood method based on the model of Tamura and Nei, within the program MEGA v6 [41]. The tree with the highest log likelihood is shown. Bootstrap replications (1000) were performed, which are expressed on each node as a percentage. The scale bar indicates the number of nucleotide substitutions per site. Accession numbers for references sequences are indicated. Sequences derived from this study were deposited in GenBank (Accession numbers - to be provided). Species (or clades) identified in this study are indicated on the right.
Theileria qPCR
The analytical specificity and sensitivity of the T. orientalis multiplex UIC assay has been reported previously [36]. The analytical specificity of the B assay was confirmed using both plasmids containing MPSP genes and Buffeli positive and negative EDTA blood extracts (data not shown). The limit of detection of the Universal, Ikeda and Chitose components of the UIC assay are 17 (8–45), 27 (8–90) and 20 (6–65) GC/μL respectively [36]. The limit of detection of the B assay used in this study was found to be similar at 20 (9–43) GC/μL. Samples yielding amplification above these levels are reported as positive (Table 1). Amongst the ticks examined, only H. longicornis was found to be positive for theilerial DNA and these were collected from cattle in the districts of Won Wron, Yarram, Bairnsdale and Warragul (Figure 1), all areas in which clinical outbreaks of theileriosis have been recently reported [37,38]. In 2013, a serological and haematological survey found evidence of theilerial infection in 10 of 15 herds from these regions of Gippsland [36]. As shown in Table 1, ticks sourced from these regions were found to harbour all 3 genotypes of T. orientalis examined (Ikeda, Chitose and Buffeli). The Ikeda genotype, which has caused clinical outbreaks in Australia [6,9,38] was found in all pools of ticks testing positive for T. orientalis. The Buffeli and Chitose genotypes were found in all, and all but two, of the T. orientalis-positive pools respectively. These results are consistent with prior studies on blood samples collected from cattle in Eastern Victoria, in which all three of these genotypes were variably detected in disparate herds [37,38]. Quantitative data indicated that parasite load was highest in ticks sourced from Won Wron and Yarram and in most cases, the Ikeda genotype was present in higher concentrations than either the Chitose or Buffeli types (Table 1). Because pools of ticks were tested, this may equate to high levels of the Ikeda parasite in individual ticks or an increased prevalence of the Ikeda type within multiple ticks.
Several thousand mosquitoes were collected on outbreak farms or within close proximity. The mosquitoes identified include; Aedes camptorhynchus, Aedes notoscriptus, Coquillettidia linealis, Culex australicus, and Culex molestus. Of the 13 pools of mosquitoes tested, one was weakly positive for T. orientalis (41 GC/μL), with only the Buffeli genotype identified over the limit of detection of the assay. Batch testing of the remaining mosquitoes also resulted in a weak positive (20 GC/μL) for T. orientalis with no genotypes exceeding the limits of detection for the respective assays. Biting flies (Dasybasis sp.) tested (n = 28) were found to be negative for T. orientalis and all genotypes.
Discussion
The epidemiology of the T. orientalis outbreaks in south-eastern Australia is largely unknown, although outbreaks in Victoria are believed to be linked to introduction of infected cattle from New South Wales to a farm near Seymour [7], at the periphery of the known range of H. longicornis (Figure 1). Prior evidence for the transmission of T. orientalis by Haemaphysalis ticks in Australia has been contradictory, but H. longicornis ticks sourced from both Japan and Australia have been shown experimentally to transmit the Japanese T. orientalis Ikeda genotype trans-stadially [2]. Indeed, the H. longicornis barcoding sequences obtained in this study suggest that the H. longicornis populations in Eastern Victoria are relatively homogenous and are closely related to those in Asia. Interestingly, the known range of H. longicornis in Australia closely mirrors areas in which outbreaks of clinical theileriosis have occurred, including an isolated population of this species in the south west of Western Australia (Figure 1). It could be hypothesised from the prior data that H. longicornis is a likely vector for T. orientalis Ikeda in Australia and the detection of T. orientalis Ikeda DNA in ticks from various regions in Victoria supports this view. Based on previous studies, the ability of H. longicornis to transmit T. orientalis Buffeli is less certain [22]. It is noteworthy therefore, that the Buffeli genotype (as well as the Chitose genotype) was detected within H. longicornis ticks along with the Ikeda genotype. This study also revealed H. longicornis as the major tick species found on cattle in Victorian herds suffering recent clinical outbreaks, further implicating this species as a likely vector of bovine theileriosis. The location of the detected theilerial DNA within H. longicornis is the focus of continuing research. Furthermore, extensive analysis of the proportions of the T. orientalis genotypes within individual ticks would be of future interest to determine whether particular genotypes are selected during the tick phase of the parasite’s lifecycle [39].
Other Haemaphysalis and Ixodes spp. have recently been implicated in transmission of T. orientalis in the Eastern Hokkaido and Okinawa prefectures via molecular screening of tick species for the presence of the parasite [11]. Interestingly, we did not detect theilerial DNA in any other tick species or march flies examined in this study. Two pools of mosquitoes were weakly positive for T. orientalis; however because these samples were close to the limit of detection of the assay, only the Buffeli genotype was identified in one of the pools. Further research is required to confirm their role as possible mechanical transmitters, as has been demonstrated for lice in Japan [40]. Studies are currently underway to determine the volume of blood required to transmit infection, and this will give an indication whether biting arthropods could pose any risk for mechanical transmission of the disease in Australia. The number of haematophagous flies caught on the outbreak farms was too small to be conclusive, and lice should also not be ruled out as a possible vector, even though no herd in the outbreak regions investigated showed evidence of louse infestations at the time of theilerial diagnosis. An understanding of the epidemiology, including the identification of vector(s), mechanical and intermediate hosts, is essential to stem the outbreaks of this emerging disease, and is the subject of ongoing investigation.
Conclusions
This paper presents the first data identifying DNA from T. orientalis in H. longicornis from herds with recent outbreaks of clinical theileriosis. This adds to the body of international evidence for its role in the life cycle and transmission of the parasite. Further studies are in progress to demonstrate the sporozoite stage of the parasite within H. longicornis and to confirm transmission of the various T. orientalis genotypes from H. longicornis to cattle.
References
Uilenberg G, Perie NM, Spanjer AAM, Franssen FFJ. Theileria orientalis, a cosmopolitan blood parasites of cattle - demonstration of the schizont stage. Res Vet Sci. 1985;38(3):352–60.
Fujisaki K, Kawazu S, Kamio T. The taxonomy of the bovine Theileria spp. Parasitol Today. 1994;10(1):31–3.
Gubbels M-J, Hong Y, van der Weide M, Qi B, Nijman IJ, Guangyuan L, et al. Molecular characterisation of the Theileria buffeli/orientalis group. Int J Parasitol. 2000;30(8):943–52.
Cufos N, Jabbar A, de Carvalho LM, Gasser RB. Mutation scanning-based analysis of Theileria orientalis populations in cattle following an outbreak. Electrophoresis. 2012;33(13):2036–40.
McFadden AMJ, Rawdon TG, Meyer J, Makin J, Morley CM, Clough RR, et al. An outbreak of haemolytic anaemia associated with infection of Theileria orientalis in naïve cattle. N Z Vet J. 2011;59(2):79–85.
Kamau J, de Vos AJ, Playford M, Salim B, Kinyanjui P, Sugimoto C. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasites Vectors. 2011;4:22.
Islam MK, Jabbar A, Campbell BE, Cantacessi C, Gasser RB. Bovine theileriosis - an emerging problem in south-eastern Australia? Infect Genet Evol. 2011;11(8):2095–7.
Izzo MM, Poe I, Horadagoda N, De Vos AJ, House JK. Haemolytic anaemia in cattle in NSW associated with Theileria infections. Aust Vet J. 2010;88(1–2):45–51.
Eamens GJ, Bailey G, Jenkins C, Gonsalves JR. Significance of Theileria orientalis types in individual affected beef herds in New South Wales based on clinical, smear and PCR findings. Vet Parasitol. 2013;196(1–2):96–105.
Eamens GJ, Gonsalves JR, Jenkins C, Collins D, Bailey G. Theileria orientalis MPSP types in Australian cattle herds associated with outbreaks of clinical disease and their association with clinical pathology findings. Vet Parasitol. 2013;191(3–4):209–17.
Yokoyama N, Sivakumar T, Ota N, Igarashi I, Nakamura Y, Yamashina H, et al. Genetic diversity of Theileria orientalis in tick vectors detected in Hokkaido and Okinawa, Japan. Infect Genet Evol. 2012;12(8):1669–75.
Ota N, Mizuno D, Kuboki N, Igarashi I, Nakamura Y, Yamashina H, et al. Epidemiological survey of Theileria orientalis infection in grazing cattle in the eastern part of Hokkaido, Japan. J Vet Med Sci. 2009;71(7):937–44.
Sarataphan N, Kakuda T, Chansiri K, Onuma M. Survey of benign Theileria parasites of cattle and buffaloes in Thailand using allele-specific polymerase chain reaction of major piroplasm surface protein gene. J Vet Med Sci. 2003;65(1):133–5.
Khukhuu A, Lan DTB, Long PT, Ueno A, Li Y, Luo Y, et al. Molecular epidemiological survey of Theileria orientalis in Thua Thien Hue Province, Vietnam. J Vet Med Sci. 2011;73(5):701–5.
Kawazu S-i, Kamio T, Kakuda T, Terada Y, Sugimoto C, Fujisaki K. Phylogenetic relationships of the benign Theileria species in cattle and Asian buffalo based on the major piroplasm surface protein (p33/34) gene sequences. Int J Parasitol. 1999;29(4):613–8.
Seddon HR. Diseases of Domestic Animals in Australia, vol. 4. Canberra: Commonwealth of Australia Department of Health; 1952.
Kamio T, Rajamanickam C, Kawazu S, Fujisaki K. Epidemiology and pathogenicity of bovine theileriosis in Malaysia. Jarq-Japan Agric Res Q. 1990;24(3):231–4.
Liu AH, Guan GQ, Liu JL, Liu ZJ, Leblanc N, Li YQ, et al. Polymorphism analysis of Chinese Theileria sergenti using allele-specific polymerase chain reaction of the major piroplasm surface protein gene. J Parasitol. 2011;97(1):116–21.
Baek BK, Soo KB, Kim JH, Hur J, Lee BO, Jung JM, et al. Verification by polymerase chain reaction of vertical transmission of Theileria sergenti in cows. Canadian J Vet Res. 2003;67:278–82.
Thomson J. Bovine anaemia detected in Western Australia. 2013. https://www.agric.wa.gov.au/news/media-release/bovine-anaemia-detected-wa. Accessed 25 Feb 2015.
Theileriosis disease discovered in Australian cattle. 2014. http://www.thecattlesite.com/news/46773/theileriosis-disease-discovered-in-australian-cattle/. Accessed 17 Apr 2015.
Stewart NP, de Vos AJ, McGregor W, Shiels I. Haemaphysalis humerosa, not H. longicornis, is the likely vector of Theileria buffeli in Australia. Aust Vet J. 1987;64(9):280–2.
Hoogstraal H, Roberts FHS, Kohls GM, Vernon JT. Review of Haemaphysalis (Kaiseriana) longicornis Neumann (resurrected) of Australia, New Zealand, New Caledonia, Fiji, Japan, Korea, and Northeastern China and USSR, and its parthenogenetic and bisexual populations (Ixodoidea, Ixodidae). J Parasitol. 1968;54(6):1197–213.
Perera PK, Gasser RB, Jabbar A. Assessment of sequence variability in a p23 gene region within and among three genotypes of the Theileria orientalis complex from south-eastern Australia. Ticks Tick-Borne Dis. 2015;6(2):123–8.
Roberts FHS. Australian Ticks. Melbourne: Commonwealth Scientific and Industrial Research Organisation, Australia; 1970.
Besier RB, Wroth R. Discovery of the tick Haemaphysalis longicornis in Western Australia. Aust Vet J. 1985;62(6):205–6.
Cane R. Profile: Haemaphysalis longicornis Neumann, 1901. 2010. http://www.smsl.co.nz/site/southernmonitoring/files/NZB/Ha%20longicornis%20Profile.pdf. Accessed 25 Feb 2015.
Klompen H, Dobson S, Barker S. A new subfamily, Bothriocrotoninae n. subfam., for the genus Bothriocroton Keirans, King & Sharrad, 1994 status amend. (Ixodida: Ixodidae), and the synonymy of Aponomma Neumann, 1899 with Amblyomma Koch, 1844. Syst Parasitol. 2002;53(2):101–7.
Jackson J, Beveridge I, Chilton NB, Andrews RH. Distributions of the paralysis ticks Ixodes cornuatus and Ixodes holocyclus in south-eastern Australia. Aust Vet J. 2007;85(10):420–4.
Riek RF. Epidemiology and transmission of Theileria sp. of cattle in Australia. Aust Vet J. 1982;59(3):89–92.
Stewart NP, De Vos AJ, Shiels I, McGregor W. The experimental transmission of Theileria buffeli of cattle in Australia by Haemaphysalis humerosa. Aust Vet J. 1987;64(3):81–3.
Stewart NP, Devos AJ, Shiels IA, Jorgensen WK. Transmission of Theileria buffeli to cattle by Haemaphysalis bancrofti fed on artificially infected mice. Vet Parasitol. 1989;34(1–2):123–7.
Laan B, Handasyde K, Beveridge I. Occurrence of the tick Haemaphysalis bancrofti Nuttall & Warburton, 1915 in Victoria with additional data on its distribution and with scanning electron micrographs of life cycle stages. Proc Roy Soc Victoria. 2011;123:189–99.
Russell RC. A Colour Photo Atlas of Mosquitoes of Southeastern Australia. Sydney: Department of Medical Entomology University of Sydney and Westmead Hospital; 1996.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9.
Bogema DR, Deutscher AT, Fell S, Collins D, Eamens GJ, Jenkins C. Development and validation of a multiplexed hydrolysis probe qPCR assay for the detection and quantification of Theileria orientalis and differentiation of clinically-relevant subtypes. J Clin Microbiol. 2015;53(3):941–50.
Read E. Prevalence and Transmission of Theileria in Cattle in Gippsland. Melbourne: La Trobe University; 2013.
Perera PK, Gasser RB, Anderson GA, Jeffers M, Bell CM, Jabbar A. Epidemiological survey following oriental theileriosis outbreaks in Victoria, Australia, on selected cattle farms. Vet Parasitol. 2013;197(3–4):509–21.
Jenkins C, Micallef M, Alex SM, Collins D, Djordjevic SP, Bogema DR. Temporal dynamics and subpopulation analysis of Theileria orientalis genotypes in cattle. Infec Genet Evol. 2015;32:199–207. In Press.
Fujisaki K, Kamio T, Kawazu S, Shimizu S, Shimura K. Theileria sergenti - experimental transmission by the long-nosed cattle louse, Linognathus vituli. Ann Trop Med Parasitol. 1993;87(2):217–8.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26.
Acknowledgements
Dr Jade Hammer is enrolled in a Master of Veterinary Science degree at the University of Sydney. The authors would like to acknowledge the financial support from Meat and Livestock Australia and The United Dairyfarmers of Victoria. We are also very grateful to all our collectors and to those farmers who allowed trapping equipment on their properties. The type specimen of H. longicornis was kindly provided by Grant Rawlin and Mallik Malipatil.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors declare that they do not have any competing interests.
Authors’ contributions
JFH conceived the study and with DE designed the study. JFH trapped and collected all arthropods investigated, and with CJ drafted the manuscript. CJ performed the DNA barcoding and phylogenetic analysis to identify the ticks, and processed the arthropods for PCR. DB conducted qPCR testing of the ticks for Theileria orientalis and genotypes. DE assisted conceptual discussions, funding and revision of the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Hammer, J.F., Emery, D., Bogema, D.R. et al. Detection of Theileria orientalis genotypes in Haemaphysalis longicornis ticks from southern Australia. Parasites Vectors 8, 229 (2015). https://doi.org/10.1186/s13071-015-0839-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0839-9